Abstract
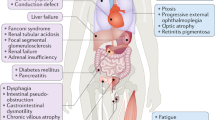
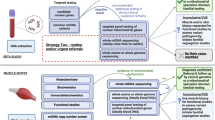
Next-generation sequencing (NGS), an innovative sequencing technology that enables the successful analysis of numerous gene sequences in a massive parallel sequencing approach, has revolutionized the field of molecular biology. Although NGS was introduced in a rather recent past, the technology has already demonstrated its potential and effectiveness in many research projects, and is now on the verge of being introduced into the diagnostic setting of routine laboratories to delineate the molecular basis of genetic disease in undiagnosed patient samples. We tested a benchtop device on retrospective genomic DNA (gDNA) samples of controls and patients with a clinical suspicion of a mitochondrial DNA disorder. This Ion Torrent Personal Genome Machine platform is a high-throughput sequencer with a fast turnaround time and reasonable running costs. We challenged the chemistry and technology with the analysis and processing of a mutational spectrum composed of samples with single-nucleotide substitutions, indels (insertions and deletions) and large single or multiple deletions, occasionally in heteroplasmy. The output data were compared with previously obtained conventional dideoxy sequencing results and the mitochondrial revised Cambridge Reference Sequence (rCRS). We were able to identify the majority of all nucleotide alterations, but three false-negative results were also encountered in the data set. At the same time, the poor performance of the PGM instrument in regions associated with homopolymeric stretches generated many false-positive miscalls demanding additional manual curation of the data.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Schon EA, DiMauro S, Hirano M : Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet 2012; 13: 878–890.
Chinnery P, Hudson G : Mitochondrial genetics. Br Med Bull 2013; 106: 135–139.
MITOMAP: A Human Mitochondrial Genome Database. Available at: http://www.mitomap.org (last accessed 15 November 2013).
Naue J, Sänger T, Schmidt U, Klein R, Lutz-Bonengel S : Factors affecting the detection and quantification of mt point heteroplasmy using Sanger sequencing and SNaPshot minisequencing. Int J Legal Med 2011; 125: 427–436.
Metzker ML : Sequencing technologies – the next generation. Nat Rev Genet 2010; 11: 31–46.
Cui H, Li F, Chen D et al: Comprehensive NGS analyses of the entire mitochondrial genome reveal new insights into the molecular diagnosis of mitochondrial DNA disorders. Genet Med 2013; 15: 388–394.
Zaragoza MV, Fass J, Diegoli M, Lin D, Arbustini E : MtDNA variant discovery and evaluation in human cardiomyopathies through NGS. PLoS One 2010; 5: e12295.
Cheng S, Higuchi R, Stoneking M : Complete mitochondrial genome amplification. Nat Genet 1994; 7: 350–351.
Li H, Handsaker B, Wysoler A et al: The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
Parson W, Strobl C, Huber G et al: Evaluation of next generation mtGenome sequencing using the Ion Torrent Personal Genome Machine (PGM). Forensic Sci Int Genet 2013; 7: 543–549.
McFarland R, Taylor RW, Chinnery PF, Howell N, Turnbull DM : A novel sporadic mutation in cytochrome c oxidase subunit II as a cause of rhabdomyolysis. Neuromuscul Disord 2004; 14: 162–166.
Loman NJ, Misra RV, Dallman TJ et al: Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 2012; 30: 434–439.
Li M, Schönberg A, Schaefer M, Schroeder R, Nasidze I, Stoneking M : Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet 2010; 87: 237–249.
He Y, Wu J, Dressman DC et al: Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 2010; 464: 610–614.
Goto H, Dickins B, Afgan E et al: A. Dynamics of mitochondrial heteroplasmy in three families investigated via a repeatable re-sequencing study. Genome Biol 2011; 12: R59.
Payne BA, Wilson IJ, Yu-Wai-Man P et al: Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet 2013; 22: 384–390.
Bragg LM, Stone G, Butler MK, Hugenholtz P, Tyson GW : Shining a light on dark sequencing: characterising errors in Ion Torrent PGM data. PLoS Comput Biol 2013; 9: e1003031.
Seneca S, Abramowicz M, Lissens W et al: A mtDNA microdeletion in a newborn girl with transient lactic acidosis. J Inherit Metab Dis 1996; 19: 115–118.
Götz A, Isohanni P, Liljeström B et al: A. Fatal neonatal lactic acidosis caused by a novel mt G7453A tRNA-Serine mutation. Pediatr Res 2012; 72: 90–94.
Santorelli FM, Tanji K, Kulikova R et al: Identification of a novel mutation in the mtDNA ND5 gene associated with MELAS. Biochem Biophys Res Commun 1997; 238: 326–328.
Goto Y, Nonaka I, Horai S : A mutation in the tRNA(Leu) gene associated with the MELAS subgroup of encephalomyopathies. Nature 1990; 348: 651.
Kirino Y, Goto Y, Campos Y, Arenas J, Suzuki T : Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc Natl Acad Sci USA 2005; 102: 7127–7132.
Acknowledgements
We thank patients and physicians for their contribution to this research project. We also want to thank S Bessems, G Van Dyck, D Daneels, B Caljon and D Croes for their contribution to this work. This project was supported by grants from the Fonds voor Wetenschappelijk Onderzoek (FWO) G.0.200, the ‘Association Belge contre les Maladies Neuro-Musculaires (ABMM)’ and the Vrije Universiteit Brussel (VUB, Brussel, Belgium) with reference nos. OZR1928 and OZRMETH3.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Seneca, S., Vancampenhout, K., Van Coster, R. et al. Analysis of the whole mitochondrial genome: translation of the Ion Torrent Personal Genome Machine system to the diagnostic bench?. Eur J Hum Genet 23, 41–48 (2015). https://doi.org/10.1038/ejhg.2014.49
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2014.49
This article is cited by
-
Analytical parameters and validation of homopolymer detection in a pyrosequencing-based next generation sequencing system
BMC Genomics (2018)
-
Assessment of mitochondrial DNA heteroplasmy detected on commercial panel using MPS system with artificial mixture samples
International Journal of Legal Medicine (2018)
-
MitoRS, a method for high throughput, sensitive, and accurate detection of mitochondrial DNA heteroplasmy
BMC Genomics (2017)
-
Accurate and comprehensive analysis of single nucleotide variants and large deletions of the human mitochondrial genome in DNA and single cells
European Journal of Human Genetics (2017)
-
Whole-mitochondrial genome sequencing in primary open-angle glaucoma using massively parallel sequencing identifies novel and known pathogenic variants
Genetics in Medicine (2015)