Abstract
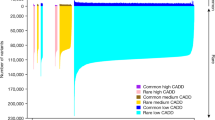
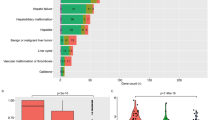
Knowledge of variant pathogenicity is key to implementing genomic medicine. We describe variability between expert reviewers in assigning pathogenicity to sequence variants in LDLR, the causal gene in the majority of cases of familial hypercholesterolemia. LDLR was sequenced on the Illumina HiSeq platform (average read depth >200 × ) in 1013 Mayo Biobank participants recruited from 2012 to 2013. Variants with a minor allele frequency (MAF) <5% predicted to be functional or referenced in HGMD (Human Gene Mutation Database) or NCBI-ClinVar databases were reviewed. To assign pathogenicity, variant frequency in population data sets, computational predictions, reported observations and patient-level data including electronic health record-based post hoc phenotyping were leveraged. Of 178 LDLR variants passing quality control, 25 were selected for independent review using either an in-house protocol or a disease/gene-specific semi-quantitative framework based on the American College of Medical Genetics and Genomics-recommended lines of evidence. NCBI-ClinVar included interpretations for all queried variants with 74% (14/19) of variants with >1 submitter showing inconsistency in classification and 26% (5/19) appearing with conflicting clinical actionability. The discordance rate (one-step level of disagreement out of five classes in variant interpretation) between the reviewers was 40% (10/25). Two LDLR variants were independently deemed clinically actionable and returnable. Interpretation of LDLR variants was often discordant among ClinVar submitters and between expert reviewers. A quantitative approach based on strength of each predefined criterion in the context of specific genes and phenotypes may yield greater consistency between different reviewers.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Fabsitz RR, McGuire A, Sharp RR et al: Ethical and practical guidelines for reporting genetic research results to study participants: updated guidelines from a National Heart, Lung, and Blood Institute working group. Circ Cardiovasc Genet 2010; 3: 574–580.
MacArthur DG, Manolio TA, Dimmock DP et al: Guidelines for investigating causality of sequence variants in human disease. Nature 2014; 508: 469–476.
Green RC, Berg JS, Grody WW et al: ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013; 15: 565–574.
Kullo IJ, Haddad R, Prows CA et al: Return of results in the genomic medicine projects of the eMERGE network. Front Genet 2014; 5: 50.
Van Driest SL, Wells QS, Stallings S et al: Association of arrhythmia-related genetic variants with phenotypes documented in electronic medical records. JAMA 2016; 315: 47–57.
Safarova MS, Kullo IJ : My Approach to the patient with familial hypercholesterolemia. Mayo Clin Proc 2016; 91: 770–786.
Sjouke B, Kusters DM, Kindt I et al: Homozygous autosomal dominant hypercholesterolaemia in the Netherlands: prevalence, genotype-phenotype relationship, and clinical outcome. Eur Heart J 2015; 36: 560–565.
Richards CS, Bale S, Bellissimo DB et al: ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med 2008; 10: 294–300.
Futema M, Plagnol V, Li K et al: Whole exome sequencing of familial hypercholesterolaemia patients negative for LDLR/APOB/PCSK9 mutations. J Med Genet 2014; 51: 537–544.
Lawrence L, Sincan M, Markello T et al: The implications of familial incidental findings from exome sequencing: the NIH Undiagnosed Diseases Program experience. Genet Med 2014; 16: 741–750.
Amendola LM, Dorschner MO, Robertson PD et al: Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res 2015; 25: 305–315.
Jurgens J, Ling H, Hetrick K et al: Assessment of incidental findings in 232 whole-exome sequences from the Baylor-Hopkins Center for Mendelian Genomics. Genet Med 2015; 17: 782–788.
Amendola LM, Jarvik GP, Leo MC et al: Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet 2016; 98: 1067–1076.
Rasmussen-Torvik LJ, Stallings SC, Gordon AS et al: Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther 2014; 96: 482–489.
Bielinski SJ, Olson JE, Pathak J et al: Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc 2014; 89: 25–33.
Gordon AS, Fulton RS, Qin X, Mardis ER, Nickerson DA, Scherer S : PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenet Genomics 2016, e-pub ahead of print 5 January 2016 doi:10.1097/FPC.0000000000000202.
Cingolani P, Platts A, Wang le L et al: A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012; 6: 80–92.
McKenna A, Hanna M, Banks E et al: The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303.
World Health OrganizationFamilial hypercholesterolemia: report of a second WHO Consultation. World Health Organization: Geneva, Switzerland, 1999 WHO publication No. WHO/HGN/FH/CONS/99.2.
Haralambos K, Whatley SD, Edwards R et al: Clinical experience of scoring criteria for Familial Hypercholesterolaemia (FH) genetic testing in Wales. Atherosclerosis 2015; 240: 190–196.
Rehm HL, Bale SJ, Bayrak-Toydemir P et al: ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013; 15: 733–747.
Hunter JE, Irving SA, Biesecker LG et al: A standardized, evidence-based protocol to assess clinical actionability of genetic disorders associated with genomic variation. Genet Med 2016; 18: 1258–1268.
Richards S, Aziz N, Bale S et al: Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424.
Dorschner MO, Amendola LM, Turner EH et al: Actionable, pathogenic incidental findings in 1000 participants' exomes. Am J Hum Genet 2013; 93: 631–640.
Johnston JJ, Rubinstein WS, Facio FM et al: Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet 2012; 91: 97–108.
Hopkins PN, Toth PP, Ballantyne CM, Rader DJ : Familial hypercholesterolemias: prevalence, genetics, diagnosis and screening recommendations from the National Lipid Association Expert Panel on Familial Hypercholesterolemia. J Clin Lipidol 2011; 5: S9–17.
Gidding SS, Ann Champagne M, de Ferranti SD et al: The agenda for familial hypercholesterolemia: a scientific statement from the American Heart Association. Circulation 2015; 132: 2167–2192.
Manrai AK, Funke BH, Rehm HL et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med 2016; 375: 655–665.
Lek M, Karczewski KJ, Minikel EV et al: Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–291.
Acknowledgements
We thank the RIGHT Protocol and Mayo Clinic Biobank study investigators, field staff, and study participants. We thank Luanne F Wussow for assistance in preparation of the manuscript. We appreciate thoughtful comments on this contribution and valuable discussions with Xiao Fan PhD (Cardiovascular Biomarkers Research Laboratory, Mayo Clinic, Rochester, MN, USA). Dr Safarova is supported by AHA Postdoctoral Fellowship Award 16POST27280004. This study was funded as part of the National Human Genome Research Institute’s electronic Medical Records and Genomics Network grants to Mayo Clinic (HG04599 and HG006379), R01 GM28157, U01 HG005137, R01 CA138461, R01 AG034676 (The Rochester Epidemiology Project), and the Mayo Clinic Center for Individualized Medicine. The sequencing platform was developed by the next-generation sequencing centers of the Pharmacogenomics Research Network supported by NIH grants U19GM61388, U19HLO69757 and U01GMO97119. The National Human Genome Research Institute and American Heart Association had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review or approval of the manuscript, including the decision to submit the manuscript for publication. Discussion of this paper by Drs Kullo and Safarova is available in the Supplementary Video.
Web resources
NCBI-ClinVar database, http://www.ncbi.nlm.nih.gov/clinvar first assessed July 2015, last assessed August 2016, LDLR Leiden Open Variation, LOVD; versions 1.1.0 build 12, 2.0 build 36, and 3.0 build 13; http://www.ucl.ac.uk/ldlr/LOVDv.1.1.0/index.php?select_db=LDLR, http://www.ucl.ac.uk/ldlr, https://grenada.lumc.nl/LOVD2/UCL-Heart/home.php?select_db=LDLR, http://databases.lovd.nl/whole_genome/genes/LDLR assessed December 2015, NHLBI-Exome Variant Server, EVS; http://evs.gs.washington.edu/EVS/ assessed December 2015.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
A supplementary video accompanies this article on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Safarova, M., Klee, E., Baudhuin, L. et al. Variability in assigning pathogenicity to incidental findings: insights from LDLR sequence linked to the electronic health record in 1013 individuals. Eur J Hum Genet 25, 410–415 (2017). https://doi.org/10.1038/ejhg.2016.193
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2016.193
This article is cited by
-
Challenging interpretation of germline TP53 variants based on the experience of a national comprehensive cancer centre
Scientific Reports (2023)
-
Familial hypercholesterolaemia: evolving knowledge for designing adaptive models of care
Nature Reviews Cardiology (2020)
-
Challenges in returning results in a genomic medicine implementation study: the Return of Actionable Variants Empirical (RAVE) study
npj Genomic Medicine (2020)
-
The complex molecular genetics of familial hypercholesterolaemia
Nature Reviews Cardiology (2019)
-
Genomic medicine for kidney disease
Nature Reviews Nephrology (2018)