Abstract
Purpose:
This article demonstrates a prominent noninvasive prenatal approach to assist the clinical diagnosis of a single-gene disorder disease, maple syrup urine disease, using targeted sequencing knowledge from the affected family.
Methods:
The method reported here combines novel mutant discovery in known genes by targeted massively parallel sequencing with noninvasive prenatal testing.
Results:
By applying this new strategy, we successfully revealed novel mutations in the gene BCKDHA (Ex2_4dup and c.392A>G) in this Chinese family and developed a prenatal haplotype-assisted approach to noninvasively detect the genotype of the fetus (transmitted from both parents).
Conclusion:
This is the first report of integration of targeted sequencing and noninvasive prenatal testing into clinical practice. Our study has demonstrated that this massively parallel sequencing–based strategy can potentially be used for single-gene disorder diagnosis in the future.
Genet Med 16 8, 594–600.
Similar content being viewed by others
Introduction
To date, 7,385 Mendelian inheritance diseases have been reported worldwide in OMIM. Single-gene disorders are genetic conditions caused by alterations or mutations primarily at one specific gene. In total, all single-gene disorders contribute to ~20% of infant mortality and ~10% of pediatric hospitalizations.1
The recent advancement in molecular genetic approaches, such as Sanger sequencing and real-time polymerase chain reaction (PCR), has allowed identification of single-gene mutations. Although these methods are generally accurate and fast, the limitation is that they focus on only a very few known mutations of known genes. On the other hand, massively parallel sequencing (MPS) has made it possible to discover novel mutations in known or unknown genes. The rapid development of this technology in the past 3 years has demonstrated great promise in clinical practice with its high efficacy and sensitivity. The identification of new genes or new mutations in single-gene disorders comes at an acceptable turnaround time and price. The new mutations of known genes reported in family studies have cumulatively expanded our genetic knowledge and therefore increased the sensitivity of prenatal tests in clinical practice.
Another hot spot in this field is the clinical application of noninvasive prenatal testing (NIPT) for fetal aneuploidies based on MPS. However, to date, only a few prenatal tests have been recommended in hospitals for some high-risk populations, for example, the test for thalassemia in the south of China. In previous studies, maternal plasma DNA sequencing has been proven to be feasible for noninvasive prenatal detection of fetal chromosome aneuploidy.2 A few studies have reported the feasibility of detecting monogenic diseases noninvasively based on MPS, but most focused on sex-linked disorders and autosomal dominant monogenic diseases. Among them, only very few studies showed the feasibility of detecting maternal inherited mutations.3
In this study, we developed a new strategy, which combines noninvasive prenatal detection of monogenic diseases with a haplotype-based method, and applied it in the case of a Chinese family with maple syrup urine disease (MSUD). For the first time, in this unexplained clinical case, we used targeted sequencing with known genes and noninvasive prenatal detection with information on the family trio (father, mother, and proband), providing a one-step solution for NIPT of a single-gene disorder.
Materials and Methods
Patient information and genetic testing strategy
The proband ( Figure 1a ), a girl, was born in February 2010. When she was 6 months old, abnormal manifestations such as fixed eyeballs and myotonia were noticed, and her brain magnetic resonance imaging scan and electroencephalogram were abnormal. Then she was diagnosed with MSUD, a type of metabolic single-gene disorder,4 by tandem mass spectrometry examination of dried blood spots and gas chromatography/mass spectrometry tests of the urine specimen.
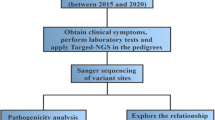
The parents (both are biological parents of the proband) are apparently healthy, as confirmed by general physical examination, and are asymptomatic for MSUD. In May 2012, the mother was pregnant again and approached the Chinese PLA General Hospital for a routine prenatal examination and expressed strong desire to know whether the fetus was healthy, preferably by a noninvasive approach. After careful genetic counseling and evaluation, a testing strategy was suggested for this family ( Figure 1a ), in which a targeted sequencing test was applied to the parents and the proband to identify the causative variation, and in parallel, NIPT using maternal cell-free DNA was performed as a screening test, as shown in Figure 1b . The finding was then confirmed at the Chinese PLA General Hospital by invasive prenatal diagnosis of the amniotic fluid (AF).
Mutation screening in MSUD-causing genes by targeted MPS
Potential mutations of the proband and her parents were analyzed using a customized capture array (NimbleGen, Madison, WI) and Illumina HiSeq2000 sequencers (Illumina, San Diego, CA). The sensitivity and efficiency of this array in MPS have been evaluated previously.5 The target region includes exons and adjacent flanking parts (total: 5.5 kb) of four known MSUD-related genes, including BCKDHA, BCKDHB, DBT, and DLD.
Peripheral blood of the parents (5 ml) and the proband (2 ml) were collected into EDTA-containing tubes when the mother was at the 13th gestational week. Peripheral blood of a healthy male adult Chinese volunteer (C1) was used as a control. All of the samples were shipped in dry ice to the BGI’s Clinical Laboratory within a few days. The genomic DNA was extracted (QIAamp DNA Blood Midi Kit, Qiagen, Hilden, Germany) and 1 μg of genomic DNA was used for each sample. Library pooling, hybridization, sequencing, and reads alignment were performed as previously described.5,6,7 Single-nucleotide variants and indels (insertion–deletion sites) were detected using the SOAPsnp software and Samtools, respectively.
Confirmation of novel mutations
To verify the data obtained by targeted MPS, real-time PCR (7500 Real-time PCR system, Applied Biosystems) and Sanger sequencing (ABI 3730 DNA Analyzer; Applied Biosystems) were performed for samples of all family members.
Genetic NIPT for fetal MSUD
Maternal blood was collected at the 13th gestational week, and 5 ml of maternal plasma was isolated by two-step centrifugation (Magnetic Genomic DNA Kit, Tiangen, Beijing, China).8 Genomic DNA from both biological parents and the proband was obtained from the previous step used in the target MPS process. Plasma DNA was obtained from 600 μl maternal plasma (Magnetic Genomic DNA Kit, Tiangen). A customer-designed NimbleGen EZ array, which targets a 181.37-Mb region of the whole exome, 1 million tag single-nucleotide polymorphisms (SNPs), and the whole major histocompatibility complex region (defined as an all-in-one array), was used for the NIPT. For each sample, 1 μg of library DNA was hybridized to 4.5 μg of mixed all-in-one baits; captured targets were then amplified and paired-end 90-bp sequencing was performed on the HiSeq2000 platform.
The SOAP2 software was used to align all the paired-end reads against the reference genome (HG19, build 37) using the parameters that included, for example, DNA segment size and aligned mismatches (-l 40 -v 5 -r 1 -s 40; l INT, for long reads with high error rate at 3'-end, those can't align whole length, then first align 5' INT bp subsequence as a seed; v INT, totally allowed mismatches in one read; r INT, how to report repeat hits, 0 = none, 1 = random one, 2 = all; s INT, min_length after soft clip). Unique reads with 0 or 1 mismatch were used for subsequent analysis after removing the duplicate reads. After filtering low-quality reads with a coverage of <4 and a value of Q20 <90%, SNP calling was performed using the SOAPsnp software in the target region. The SNPs that were homozygous in both parents but of different types were selected to calculate the fetal concentration in maternal plasma according to the formula:

where ε stands for the cell-free fetal DNA concentration in maternal plasma and dfather and dmother stand for the depth of the special base of the father and mother, respectively.
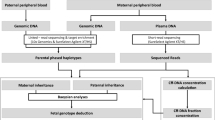
A haplotype-assisted method based on MPS was introduced by the NIPT for MSUD in the fetus. Briefly, parental haplotypes were constructed using genomic information from the data on the family trio(Supplementary Materials and Methods online). The fetal genotype had four possible combinations inheritable from the parents. The probability of each mixed state of maternal genotype and different fetal combinations was calculated for each SNP marker in maternal plasma. Finally, according to the linkage relationship obtained from the parental haplotype, we established a Hidden Markov Model with consideration of the state of SNP markers and the parental recombination information. By this strategy, the fetal genotype and haplotype can be derived in a single step (details in Supplementary Materials and Methods online).
Prenatal diagnosis using AF obtained through invasive procedure
AF obtained through an invasive procedure at the 19th gestational week was used for prenatal diagnostic genetic testing. After DNA extraction, real-time PCR and Sanger sequencing for target variations were performed as described above.
Results
Workflow
Figure 1c summarizes the workflow of this study. In the first trimester, pretest counseling for the pregnant mother was performed by senior obstetricians at the Department of Obstetrics and Gynecology. The detailed recommendations were as follows: (i) All mutations identified by the targeted MPS of the proband must be validated by other available techniques, for example, Sanger sequencing or real-time PCR; (ii) The NIPT test is mostly intended as a pilot study for the noninvasive prenatal detection of known mutations identified in step i at the gestational age of 12–24 weeks. The result is supplementary to the results from invasive prenatal testing; (iii) clinical decision will be made on the result of invasive prenatal testing; (iv) this testing strategy cannot be applied if no mutations are identified by the targeted MPS for the proband at step i; (v) this test is not intended to identify new genes in unexplained cases; (vi) all molecular tests and procedures were performed at the BGI, Shenzhen, in a clinical laboratory certified by the International Organization for Standardization/International Electrotechnical Commission 17025 and accredited by the Ministry of Health. The total turnaround time ranged from 6 to 7 weeks.
Mutation identification by targeted MPS and validation
We sequenced the target regions for the family and generated a mean depth of 180-fold per family member, including both parents and the proband. Approximately 99.94% of the targeted bases were sufficiently covered ( Table 1 ). Figure 2 shows the average depth for the BCKDHA gene of the four samples tested by MPS: the average depth for the gene BCKDHA is homogeneous in each sample, and the average sequencing coverage is more than 99%, showing high experimental quality.
Genetic analysis of the family. (a) The average depth plot of the family. Rectangle shows the average sequencing depth for all nine exons in the BCKDHA gene of the family. (b) Sanger sequencing confirmation of the missense mutation identified by targeted DNA-HiSeq. (c) quantitative real-time polymerase chain reaction (PCR) analysis. Relative amplification value calculated from the data of quantitative real-time PCRs for detecting possible duplications in BCKDHA exons 2–4 of the family. (d) Structural bases of novel missense mutations in E1a (p.Y131C). The mutant region is shown in white.
The sequencing depth of exons 2–4 of the gene BCKDHA is ~50% higher in the proband (II:1) and her father (I:1), indicating a high incidence of heterozygous duplication of these three exons ( Figure 2a ). To address whether the high read depth at exons 2–4 in II:1 and I:1 was due to capture or sequencing bias, we analyzed the depth of BCKDHA exons 2–4 in 25 human individual control samples performed in the same lane (data not shown); no duplication was detected, suggesting that II:1 and I:1 contain duplication of exons 2–4 of the BCKDHA gene that may be associated with MSUD.
All single-nucleotide variants and indels were determined according to the criteria described before.5 All led to a potential mutation, c.392A>G in the gene BCKDHA. Sanger sequencing validated that the suspected novel missense mutation (c.392A>G) exists in the proband and her mother ( Figure 2b ), consistent with the MPS results. We then used the SIFT and PolyPhen2 (Polymorphism Phenotyping v2) tools to assess the effects of the missense mutation on protein function, and this mutation is predicted to be detrimental. Evolutionary conservation analysis revealed that the novel missense mutation was highly conserved across species (data not shown). Molecular modeling of the c.392A>G mutation (p.Y131C) with PyMOL software is shown in Figure 2d , indicating that it would induce local secondary structure changes.
Real-time PCR further confirmed the heterozygous duplication of exons 2–4 in the BCKDHA gene of the proband (II:1) and her father (I:1). A total of three primer pairs (Supplementary Table S1 online) were designed targeting BCKDHA exons 2–4. The results are shown in Figure 2c : the relative amplification levels of exons 2–4 in the proband were 1.60, 1.44, and 1.45, respectively, ~150% of the relative amplification level in C1, suggesting that there were heterozygous duplications of exons 2–4 in the BCKDHA gene of the proband, who is a carrier of this variation. The relative amplification value in the proband’s father also indicated heterozygous duplications of exons 2–4 in one allele of the BCKDHA gene ( Figure 2c ). There were significant differences between II:1 and C1, as well as between I:1 and C1 (P < 0.01). The relative amplification level in the proband’s mother was normal. The real-time PCR results were also consistent with those of the target-region MPS.
Genetic NIPT for MSUD
After basic data processing, 36.89 Mb, 46.47 Mb, and 34.07 Mb clean mapped reads of the target regions of genomes of the mother, the father, and the proband, respectively, were obtained, and the mean depths of the target region on chromosome 19 were 18.03, 22.10, and 16.86, respectively. For maternal plasma DNA, a total of 2.50M clean reads were acquired, and the coverage depth was 39.52 on chromosome 19. After the SNP-calling process by SOAPsnp, a total of 7,466, 9,027, and 7,267 candidate SNP markers in the target region of chromosome 19 in mother, father, and proband were contributed for phasing ( Table 2 ).
With the makers of the proband and her parents, we recovered the parental haplotype of chromosome 19 in 6,148 loci based on Mendel’s laws. And using a sensitive Hidden Markov Model, we identified the parental transmitted alleles and recombination break points in 5,480 loci of maternal plasma, assisted by the parental haplotype information (Supplementary Materials and Methods online). In addition, a one-time recombination of chromosome 19 in the maternal and paternal allele was identified, and the break points were both located in the short arm ( Figure 3 ). Our results showed that in the causative gene, BCKDHA, the fetus inherited the same alleles as that of the proband, which indicated it was also an MSUD patient.
Noninvasive prenatal testing using maternal plasma DNA by massively parallel sequencing (MPS). (a) Flowchart of noninvasive test pipeline; (b) schematic diagram of the trio phasing and haplotype recovery; (c) the probability of fetal inherited haplotype. x-Axis represents the loci on chromosome 19, and y-axis represents the logarithmic values of the odds ratios in different combinations of the fetal haplotype. The blue line represents the paternal allele, and the red line represents the maternal allele. The higher the value, the greater the probability that the fetus inherited the haplotype 1.
Prenatal diagnosis using AF sample
Amniocentesis was performed at the 19th week of gestational age, and 30 ml of AF was obtained. Real-time PCR and Sanger sequencing were performed on the AF DNA. The result showed the fetus inherited both the heterozygous exons 2–4 duplication and the novel missense mutation (c.392A>G) in the BCKDHA gene, the same as the proband. DNA testing found no evidence of significant maternal cell contamination (data not shown).
Posttest counseling and clinical outcome
Along with all the results from the invasive prenatal diagnosis and the NIPT testing by the 20th gestational week, it was concluded that the fetus was affected by the heterozygous exons 2–4 duplication and the novel missense mutation (c.392A>G) in the BCKDHA gene, the same as the proband. With careful posttest counseling, the couple finally decided to terminate the pregnancy. Muscular tissues of the aborted fetus were obtained and were validated by real-time PCR and Sanger sequencing, and the results were in accordance with that of AF DNA. The opportunity of conceiving a healthy child by in vitro fertilization and preimplantation genetic diagnosis has been presented to the couple and prenatal testing is strongly recommended for the next pregnancy.
Discussion
In the past 5 years, with the developments in targeted sequencing by MPS technology, accurate detection of mutations in single-gene disorders has been made available. With acceptable cost and reasonable turnaround time, this new technology has advanced from laboratory research to clinical application. However, prenatal diagnosis of single-gene disorders creates more challenges regarding the accuracy of technology and ethical implications.
The discovery of cell-free fetal DNA in maternal plasma has provided a noninvasive approach for the prenatal genetic testing of fetal aneuploidies and single-gene disorders. NIPT using MPS to detect fetal common chromosomal aneuploidies from maternal plasma samples has been validated and introduced into clinical practice in several regions.2,9,10 However, investigation for the assessment of single-gene disorders by NIPT is still limited in practice and is difficult, mainly because the background of maternal genomic DNA greatly affects the accuracy of the fetal genetic test. So far, only a few studies have been reported, especially for autosomal recessive diseases such as MSUD.
Here, we developed a new method that identifies new variations of known genes and then diagnoses single-gene disorders by a noninvasive approach; this is the first report combining targeted MPS and NIPT in clinical practice.
Targeted MPS was first performed for a Chinese family of three who were suspected to have an MSUD child, allowing identification of potential disease-causing variations in both parents and the proband. Heterozygosity for new MSUD variations in the BCKDHA gene was identified in the proband: a novel large duplication (Ex2_4dup), which came from the father, who was heterozygous at this locus, and a novel c.392A>G missense mutation, which came from the mother, who was also heterozygous at this locus. The two mutations identified have not been reported before, and to our knowledge, this is the first report of a large duplication in the gene BCKDHA associated with MSUD. The two novel mutations are expected to affect the function of E1a (E1-alpha, subunit of the branched-chain α-ketoacid dehydrogenase complex).11 E1a-129 is recognized as a polypeptide binding site in the tetramer interface; the novel p.Y131C mutation identified is close to E1a-129 and may have a negative effect on the structural integrity of the protein by disrupting tetramer assembly.
Second, for prenatal genetic testing, we constructed the parental haplotype based on the sequencing data of the family trio. Previously, we developed a fetal whole-genome recovery method using genomics information of grandparents, with an accuracy of 98.57% and 95.37% for the paternal and maternal autosomal alleles, respectively. This proof-of-concept strategy has high requirements for genomic samples from family members of three generations, and experimental techniques and analysis are rather complicated to be used in a clinical setting. Meanwhile, in most clinical cases in China, grandparents’ information and samples are not readily available, and most parents come for medical assistance because they already have children with symptoms of some suspected genetic disorder, more often undiagnosed. Therefore, compared with previous studies on parental haplotype construction, which required genomic information of four grandparents’ samples and a complicated experimental procedure,12,13,14 the strategy developed in this study used only genomic information from the two parents, the proband, and maternal plasma, which is more feasible in clinical applications.
Last, combined with sequencing data of the maternal plasma, fetal haplotype was constructed and mutations inherited from both parents were identified with high accuracy. Another advantage is that this strategy is not affected by fetal DNA fraction in maternal plasma; therefore, this can be a promising workflow for single-gene disorder diagnosis in future. Using noninvasive testing as a reference, a final decision was made at the gestational age of the 25th week based on the results of invasive testing.
Turnaround time and cost are issues and challenges for MPS-based prenatal testing. In this work, the whole testing procedure was completed in 50 calendar days and diagnosis was reached at the gestational age of the 20th week. With the rapid development of an MPS-based test, the reporting time could be reduced to 3 days or less.15,16 Moreover, we believe the cost will decrease to similar levels as conventional invasive diagnosis methods with the advances in high-throughput sequencing.
With the progression of sequencing and NIPT technology, noninvasive prenatal diagnosis has shown enormous potential in single-gene disorders. In the coming years, with growing clinical needs, it is very likely that prenatal diagnosis will go from invasive to noninvasive approaches, and the current NIPT testing spectrum will expand from chromosomal abnormality detection to diagnosis of single-gene disorders with a higher sensitivity. This is a proof-of-concept work showing the potential usage of targeted MPS and haplotype-assisted methodology for a clinical case. More data are needed for a full evaluation of this technology to incorporate it into clinical practice.
Disclosure
The authors declare no conflict of interest.
References
Bell CJ, Dinwiddie DL, Miller NA, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med 2011;3:65ra4.
Dan S, Wang W, Ren J, et al. Clinical application of massively parallel sequencing-based prenatal noninvasive fetal trisomy test for trisomies 21 and 18 in 11,105 pregnancies with mixed risk factors. Prenat Diagn 2012;32:1225–1232.
Lo YM, Chan KC, Sun H, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med 2010;2:61ra91.
Nellis MM, Kasinski A, Carlson M, et al. Relationship of causative genetic mutations in maple syrup urine disease with their clinical expression. Mol Genet Metab 2003;80:189–195.
Wei X, Ju X, Yi X, et al. Identification of sequence variants in genetic disease-causing genes using targeted next-generation sequencing. PLoS ONE 2011;6:e29500.
Wei X, Sun Y, Xie J, et al. Next-generation sequencing identifies a novel compound heterozygous mutation in MYO7A in a Chinese patient with Usher Syndrome 1B. Clin Chim Acta 2012;413:1866–1871.
Sun Y, Wang L, Wei X, et al. Analysis of a Chinese pedigree with Zellweger syndrome reveals a novel PEX1 mutation by next-generation sequencing. Clin Chim Acta 2013;417:57–61.
Lau TK, Chen F, Pan X, et al. Noninvasive prenatal diagnosis of common fetal chromosomal aneuploidies by maternal plasma DNA sequencing. J Matern Fetal Neonatal Med 2012;25:1370–1374.
Chiu RW, Chan KC, Gao Y, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA 2008;105:20458–20463.
Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR . Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA 2008;105:16266–16271.
Pettit FH, Yeaman SJ, Reed LJ . Purification and characterization of branched chain alpha-keto acid dehydrogenase complex of bovine kidney. Proc Natl Acad Sci USA 1978;75:4881–4885.
Chen S, Ge H, Wang X, et al. Haplotype-assisted accurate non-invasive fetal whole genome recovery through maternal plasma sequencing. Genome Med 2013;5:18.
Kitzman JO, Snyder MW, Ventura M, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med 2012;4:137ra76.
Fan HC, Gu W, Wang J, Blumenfeld YJ, El-Sayed YY, Quake SR . Non-invasive prenatal measurement of the fetal genome. Nature 2012;487:320–324.
Phimister EG, Feero WG, Guttmacher AE . Realizing genomic medicine. N Engl J Med 2012;366:757–759.
Rothberg JM, Hinz W, Rearick TM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011;475:348–352.
Acknowledgements
We thank all the blood donors for their invaluable contributions to this study. We thank our colleagues at the BGI for help in sequencing. We thank Jia Sophie Lu from Complete Genomics for helping us to revise the manuscript. The study received financial support from the Shenzhen Development and Reform Commission project, the Guangdong Provincial Science and Technology Department Planning Project, and the Shenzhen Birth Defect Screening Project Lab (JZF no. (2011) 861). This research was also supported by grants from the Tianjin Binhai New Area Science and Technology Commission (no. 2011-BK120011). Experiments for this study were performed in China and comply with the current laws of the country. Ethics approval was obtained from the ethics committee of BGI, Shenzhen.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Table S1
(DOC 35 kb)
Supplementary Materials and Methods
(DOC 628 kb)
Rights and permissions
About this article
Cite this article
You, Y., Sun, Y., Li, X. et al. Integration of targeted sequencing and NIPT into clinical practice in a Chinese family with maple syrup urine disease. Genet Med 16, 594–600 (2014). https://doi.org/10.1038/gim.2013.197
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/gim.2013.197
Keywords
This article is cited by
-
Noninvasive prenatal diagnosis of monogenic disorders based on direct haplotype phasing through targeted linked-read sequencing
BMC Medical Genomics (2021)
-
Non-invasive prenatal sequencing for multiple Mendelian monogenic disorders using circulating cell-free fetal DNA
Nature Medicine (2019)
-
Haplotype-based Noninvasive Prenatal Diagnosis of Hyperphenylalaninemia through Targeted Sequencing of Maternal Plasma
Scientific Reports (2018)
-
Noninvasive prenatal diagnosis of 21-Hydroxylase deficiency using target capture sequencing of maternal plasma DNA
Scientific Reports (2017)
-
Attitudes and Knowledge of Maternal‐Fetal Medicine Fellows Regarding Noninvasive Prenatal Testing
Journal of Genetic Counseling (2016)