Abstract
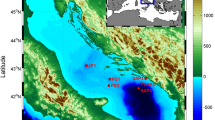
It has been long debated as to whether marine microorganisms have a ubiquitous distribution or patterns of biogeography, but recently a consensus for the existence of microbial biogeography is emerging. However, the factors controlling the distribution of marine bacteria remain poorly understood. In this study, we combine pyrosequencing and traditional Sanger sequencing of the 16S rRNA gene to describe in detail bacterial communities from the deep Arctic Ocean. We targeted three separate water masses, from three oceanic basins and show that bacteria in the Arctic Ocean have a biogeography. The biogeographical distribution of bacteria was explained by the hydrography of the Arctic Ocean and subsequent circulation of its water masses. Overall, this first taxonomic description of deep Arctic bacteria communities revealed an abundant presence of SAR11 (Alphaproteobacteria), SAR406, SAR202 (Chloroflexi) and SAR324 (Deltaproteobacteria) clusters. Within each cluster, the abundance of specific phylotypes significantly varied among water masses. Water masses probably act as physical barriers limiting the dispersal and controlling the diversity of bacteria in the ocean. Consequently, marine microbial biogeography involves more than geographical distances, as it is also dynamically associated with oceanic processes. Our ocean scale study suggests that it is essential to consider the coupling between microbial and physical oceanography to fully understand the diversity and function of marine microbes.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Agogue H, Brink M, Dinasquet J, Herndl GJ . (2008). Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature 456: 788–791.
Azam F, Malfatti F . (2007). Microbial structuring of marine ecosystems. Nat Rev Micro 5: 782–791.
Bano N, Hollibaugh JT . (2002). Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl Environ Microbiol 68: 505–518.
Broecker WS . (1991). The great ocean conveyor. Oceanography 4: 79–89.
Broecker WS . (1997). Thermohaline circulation, the achilles heel of our climate system: will man-made CO2 upset the current balance? Science 278: 1582–1588.
Darling KF, Kucera M, Pudsey CJ, Wade CM . (2004). Molecular evidence links cryptic diversification in polar planktonic protists to Quaternary climate dynamics. Proc Natl Acad Sci USA 101: 7657–7662.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU et al. (2006). Community genomics among stratified microbial assemblages in the ocean's interior. Science 311: 496–503.
Dethlefsen L, Huse S, Sogin ML, Relman DA . (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6: e280.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Felsenstein J . (2004). PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author Department of Genome Sciences, University of Washington: Seattle, USA.
Field KG, Gordon D, Wright T, Rappe M, Urback E, Vergin K et al. (1997). Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol 63: 63–70.
Fuhrman JA, Davis AA . (1997). Widespread Archaea and novel Bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar Ecol Prog Ser 150: 275–285.
Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL et al. (2008). A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci 105: 7774–7778.
Galand PE, Lovejoy C, Pouliot J, Garneau ME, Vincent WF . (2008). Microbial community diversity and heterotrophic production in a coastal Arctic ecosystem: a Stamukhi lake and its source waters. Limnol Oceanogr 53: 813–823.
Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C . (2009a). Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J 3: 860–869.
Galand PE, Lovejoy C, Hamilton AK, Ingram RG, Pedneault E, Carmack E . (2009b). Archaeal diversity and a gene for ammonia oxidation are coupled to oceanic circulation. Environ Microbiol 11: 971–980.
Gallagher JM, Carton mW, Eardly DF, Patching JW . (2004). Spatio-temporal variability and diversity of water column prokaryotic communities in the eastern North Atlantic. FEMS Microbiol Ecol 47: 249–262.
Garcia-Martinez J, Rodriguez-Valera F . (2000). Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of Group I. Mol Ecol 9: 935–948.
Giovannoni SJ, Rappe MS, Vergin KL, Adair NL . (1996). 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the Green Non-Sulfur bacteria. Proc Natl Acad Sci USA 93: 7979–7984.
Giovannoni SJ, Stingl U . (2005). Molecular diversity and ecology of microbial plankton. Nature 437: 343–348.
Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, Cho JC et al. (2008). The small genome of an abundant coastal ocean methylotroph. Environ Microbiol 10: 1771–1782.
Gonzalez JM, Simo R, Massana R, Covert JS, Casamayor EO, Pedros-Alio C et al. (2000). Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol 66: 4237–4246.
Gordon DA, Giovannoni SJ . (1996). Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific oceans. Appl Environ Microbiol 62: 1171–1177.
Hamilton AK, Lovejoy C, Galand PE, Ingram RG . (2008). Water masses and biogeography of picoeukaryote assemblages in a cold hydrographically complex system. Limnol Oceanogr 53: 922–935.
Hammer Ø, Harper DAT, Ryan PD . (2001). PAST: Paleontological statistics software package for education and data analysis. Paleontol Electro 4: 9.
Hansman RL, Griffin S, Watson JT, Druffel ERM, Ingalls AE, Pearson A et al. (2009). The radiocarbon signature of microorganisms in the mesopelagic ocean. Proc Natl Acad Sci USA 106: 6513–6518.
Herndl GJ, Agogue H, Baltar F, Reinthaler T, Sintes E, Varela MM . (2008). Regulation of aquatic microbial processes: the ‘microbial loop’ of the sunlit surface waters and the dark ocean dissected. Aqua Microbial Ecol 53: 59–68.
Hewson I, Steele JA, Capone DG, Fuhrman JA . (2006). Remarkable heterogeneity in meso- and bathypelagic bacterioplankton assemblage composition. Limnol Oceanogr 51: 1274–1283.
Hosia A, Stemmann L, Youngbluth M . (2008). Distribution of net-collected planktonic cnidarians along the northern Mid-Atlantic Ridge and their associations with the main water masses. Deep-Sea Res II 55: 106–118.
Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA et al. (2007). Microbial population structures in the deep marine biosphere. Science 318: 97–100.
Huber T, Faulkner G, Hugenholtz P . (2004). Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20: 2317–2319.
Huse S, Huber J, Morrison H, Sogin M, Welch D . (2007). Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8: R143.
Jones EP, Rudels B, Anderson LG . (1995). Deep waters of the Arctic Ocean: origins and circulation. Deep Sea Res Pt I 42: 737–760.
Karl DM . (2002). Nutrient dynamics in the deep blue sea. Trends Microbiol 10: 410–418.
Karner MB, DeLong EF, Karl DM . (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507–510.
Konstantinidis KT, DeLong EF . (2008). Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J 2: 1052–1065.
Lavik G, Stuhrmann T, Bruchert V, Van der Plas A, Mohrholz V, Lam P et al. (2009). Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature 457: 581–584.
Lopez-Garcia P, Lopez-Lopez A, Moreira D, Rodriguez-Valera F . (2001). Diversity of free-living prokaryotes from a deep-sea site at the Antarctic Polar Front. FEMS Microbiol Ecol 36: 193–202.
Macdonald RW, Carmack EC . (1991). Age of Canada Basin deep waters: a way to estimate primary production for the Arctic Ocean. Science 254: 1348–1350.
Malmstrom RR, Straza TRA, Cottrell MT, Kirchman DL . (2007). Diversity, abundance, and biomass production of bacterial groups in the western Arctic Ocean. Aqua Microbial Ecol 47: 45–55.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380.
McLaughlin FA, Carmack EC, Macdonald RW, Melling H, Swift JH, Wheeler PA et al. (2004). The joint roles of Pacific and Atlantic-origin waters in the Canada Basin, 1997–1998. Deep Sea Res Pt I 51: 107–128.
Morris RM, Rappe MS, Urbach E, Connon SA, Giovannoni SJ . (2004). Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl Environ Microbiol 70: 2836–2842.
Pham VD, Konstantinidis KT, Palden T, DeLong EF . (2008). Phylogenetic analyses of ribosomal DNA-containing bacterioplankton genome fragments from a 4000 m vertical profile in the North Pacific Subtropical Gyre. Environ Microbiol 10: 2313–2330.
Pommier T, Canback B, Riemann L, Bostrom KH, Simu K, Lundberg P et al. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol 16: 867–880.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Rudels B, Jones PE, Schauer U, Eriksson P . (2004). Atlantic sources of the Arctic Ocean surface and halocline waters. Polar Res 23: 181–208.
Schloss PD, Handelsman J . (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71: 1501–1506.
Sexton PF, Norris RD . (2008). Dispersal and biogeography of marine plankton: long-distance dispersal of the foraminifer Truncorotalia truncatulinoides. Geology 36: 899–902.
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the under explored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Spencer-Cervato C, Thierstein H . (1997). First appearance of Globorotalia truncatulinoides: Cladogenesis and immigration. Mar Micropaleontol 30: 267–291.
Stevens H, Ulloa O . (2008). Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ Microbiol 10: 1244–1259.
Stommel H . (1958). The abyssal circulation. Deep-Sea Res 5: 80–82.
Suzuki MT, Béjà O, Taylor LT, DeLong EF . (2001). Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ Microbiol 3: 323–331.
Tang CCL, Ross CK, Yao T, Petrie B, DeTracey BM, Dunlap E . (2004). The circulation, water masses and sea-ice of Baffin Bay. Prog Oceanogr 63: 183–228.
Teira E, Lebaron P, van Aken H, Herndl GJ . (2006). Distribution and activity of bacteria and archaea in the deep water masses of the North Atlantic. Limnol Oceanogr 51: 2131–2144.
Varela MM, van Aken HM, Herndl GJ . (2008a). Abundance and activity of Chloroflexi-type SAR202 bacterioplankton in the meso- and bathypelagic waters of the (sub)tropical Atlantic. Environ Microbiol 10: 1903–1911.
Varela MM, van Aken HM, Sintes E, Herndl GJ . (2008b). Latitudinal trends of Crenarchaeota and bacteria in the meso- and bathypelagic water masses of the Eastern North Atlantic. Environ Microbiol 10: 110–124.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Wright TD, Vergin KL, Boyd PW, Giovannoni SJ . (1997). A novel delta-subdivision proteobacterial lineage from the lower ocean surface layer. Appl Environ Microbiol 63: 1441–1448.
Acknowledgements
We deeply appreciate the assistance and support of the men and women of the Canadian Coast Guard icebreaker CCGS Louis St Laurent, and we acknowledge financial and ship time support from Fisheries and Oceans Canada and the Canadian International Polar Year Program's Canada's Three Oceans project and the Nansen and Amundsen Basins Observational System project. PE Galand was supported by a Marie Curie Grant (CRENARC MEIF-CT-2007-040247) and EO Casamayor by the Spanish Grant CGL2006-12058-BOS. C Lovejoy would like to acknowledge the support of the Natural Sciences and Engineering Council, Canada (NSERC) Special Research Opportunity Fund and ArcticNet. Deep Arctic samples were collected by K Scarcella, E Didierjean and M-É Garneau. Pyrosequencing was supported by a Keck foundation grant to M Sogin and L Ameral Zettler. This is a contribution to the International Census of Marine Microbes (ICOMM). We also thank three anonymous reviewers for suggestions and constructive comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary information
Rights and permissions
About this article
Cite this article
Galand, P., Potvin, M., Casamayor, E. et al. Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J 4, 564–576 (2010). https://doi.org/10.1038/ismej.2009.134
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2009.134
Keywords
This article is cited by
-
The distribution and diversity of eukaryotic phytoplankton in the Icelandic marine environment
Scientific Reports (2023)
-
Vertical segregation and phylogenetic characterization of archaea and archaeal ammonia monooxygenase gene in the water column of the western Arctic Ocean
Extremophiles (2023)
-
Oceanographic setting influences the prokaryotic community and metabolome in deep-sea sponges
Scientific Reports (2022)
-
Major ocean currents may shape the microbiome of the topshell Phorcus sauciatus in the NE Atlantic Ocean
Scientific Reports (2021)
-
Comparison of picoeukaryote community structures and their environmental relationships between summer and autumn in the southern Chukchi Sea
Extremophiles (2021)