Abstract
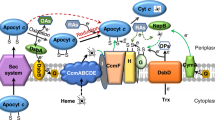
Most of the Shewanella species contain two periplasmic nitrate reductases (NAP-α and NAP-β), which is a unique feature of this genus. In the present study, the physiological function and evolutionary relationship of the two NAP systems were studied in the deep-sea bacterium Shewanella piezotolerans WP3. Both of the WP3 nap gene clusters: nap-α (napD1A1B1C) and nap-β (napD2A2B2) were shown to be involved in nitrate respiration. Phylogenetic analyses suggest that NAP-β originated earlier than NAP-α. Tetraheme cytochromes NapC and CymA were found to be the major electron deliver proteins, and CymA also served as a sole electron transporter towards nitrite reductase. Interestingly, a ΔnapA2 mutant with the single functional NAP-α system showed better growth than the wild-type strain, when grown in nitrate medium, and it had a selective advantage to the wild-type strain. On the basis of these results, we proposed the evolution direction of nitrate respiration system in Shewanella: from a single NAP-β to NAP-β and NAP-α both, followed by the evolution to a single NAP-α. Moreover, the data presented here will be very useful for the designed engineering of Shewanella for more efficient respiring capabilities for environmental bioremediation.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Balch WE, Wolfe RS . (1976). New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol 32: 781–791.
Brondijk THC, Fiegen D, Richardson DJ, Cole JA . (2002). Roles of NapF, NapG and NapH, subunits of the Escherichia coli periplasmic nitrate reductase, in ubiquinol oxidation. Mol Microbiol 44: 245–255.
Brondijk THC, Nilavongse A, Filenko N, Richardson DJ, Cole JA . (2004). NapGH components of the periplasmic nitrate reductase of Escherichia coli K-12: location, topology and physiological roles in quinol oxidation and redox balancing. Biochem J 379: 47–55.
Cruz-Garcia C, Murray AE, Klappenbach JA, Stewart V, Tiedje JM . (2007). Respiratory nitrate ammonification by Shewanella oneidensis MR-1. J Bacteriol 189: 656–662.
Drummond AJ, Aston B, Buxton S, Cheung M, Heled J, Kearse M et al. (2010). Geneious v4.8. Available from http://www.geneious.com.
Edwards RA, Keller LH, Schifferli DM . (1998). Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207: 149–157.
Gao HC, Yang ZK, Barua S, Reed SB, Romine MF, Nealson KH et al. (2009). Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. ISME J 3: 966–976.
Gao HC, Yang ZK, Wu LY, Thompson DK, Zhou JZ . (2006). Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J Bacteriol 188: 4560–4569.
Gralnick JA, Vali H, Lies DP, Newman DK . (2006). Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. P Natl Acad Sci USA 103: 4669–4674.
Gross R, Eichler R, Simon J . (2005). Site-directed modifications indicate differences in axial haem c iron ligation between the related NrfH and NapC families of multihaem c-type cytochromes. Biochem J 390: 689–693.
Huelsenbeck JP, Ronquist F . (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755.
Jepson BJ, Marietou A, Mohan S, Cole JA, Butler CS, Richardson DJ . (2006). Evolution of the soluble nitrate reductase: defining the monomeric periplasmic nitrate reductase subgroup. Biochem Soc Trans 34: 122–126.
Kern M, Simon J . (2008). Characterization of the NapGH quinol dehydrogenase complex involved in Wolinella succinogenes nitrate respiration. Mol Microbiol 69: 1137–1152.
Lenski RE, Rose MR, Simpson SC, Tadler SC . (1991). Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Naturalist 138: 1315–1341.
Maillard J, Spronk CAEM, Buchanan G, Lyall V, Richardson DJ, Palmer T et al. (2007). Structural diversity in twin-arginine signal peptide-binding proteins. P Natl Acad Sci USA 104: 15641–15646.
McGinnis S, Madden TL . (2004). BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32: W20–W25.
Miller TL, Wolin MJ . (1974). A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl Microbiol 27: 985–987.
Moreno-Vivian C, Cabello P, Martinez-Luque M, Blasco R, Castillo F . (1999). Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J Bacteriol 181: 6573–6584.
Murphy JN, Saltikov CW . (2007). The cymA gene, encoding a tetraheme c-type cytochrome, is required for arsenate respiration in Shewanella species. J Bacteriol 189: 2283–2290.
Myers JM, Myers CR . (2000). Role of the tetraheme cytochrome CymA in anaerobic electron transport in cells of Shewanella putrefaciens MR-1 with normal levels of menaquinone. J Bacteriol 182: 67–75.
Pittman MS, Elvers KT, Lee L, Jones MA, Poole RK, Park SF et al. (2007). Growth of Campylobacter jejuni on nitrate and nitrite: electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol Microbiol 63: 575–590.
Potter LC, Millington P, Griffiths L, Thomas GH, Cole JA . (1999). Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth? Biochem J 344: 77–84.
Richardson DJ, Berks BC, Russell DA, Spiro S, Taylor CJ . (2001). Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell Mol Life Sci 58: 165–178.
Sears HJ, Little PJ, Richardson DJ, Berks BC, Spiro S, Ferguson SJ . (1997). Identification of an assimilatory nitrate reductase in mutants of Paracoccus denitrificans GB17 deficient in nitrate respiration. Arch Microbiol 167: 61–66.
Simpson PJ, Richardson DJ, Codd R . (2010). The periplasmic nitrate reductase in Shewanella: the resolution, distribution and functional implications of two NAP isoforms, NapEDABC and NapDAGHB. Microbiol 156: 302–312.
Spector MP, del Portillo FG, Bearson SMD, Mahmud A, Magut M, Finlay BB et al. (1999). The rpoS-dependent starvation-stress response locus stiA encodes a nitrate reductase (narZYWV) required for carbon-starvationinducible thermotolerance and acid tolerance in Salmonella typhimurium. Microbiol-Uk 145: 3035–3045.
Stewart V, Lu Y, Darwin AJ . (2002). Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J Bacteriol 184: 1314–1323.
Stolz JF, Basu P . (2002). Evolution of nitrate reductase: molecular and structural variations on a common function. Chembiochem 3: 198–206.
Tabata A, Yamamoto I, Matsuzaki M, Satoh T . (2005). Differential regulation of periplasmic nitrate reductase gene (napKEFDABC) expression between aerobiosis and anaerobiosis with nitrate in a denitrifying phototroph Rhodobacter sphaeroides f sp denitrificans. Arch Microbiol 184: 108–116.
Thompson JD, Plewniak F, Poch O . (1999). A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res 27: 2682–2690.
Wang F, Li Q, Xiao X . (2007). A novel filamentous phage from the deep-sea bacterium Shewanella piezotolerans WP3 is induced at low temperature. J Bacteriol 189: 7151–7153.
Wang F, Xiao X, Ou HY, Gai YB, Wang FP . (2009). Role and regulation of fatty acid biosynthesis in the response of Shewanella piezotolerans WP3 to different temperatures and pressures. J Bacteriol 191: 2574–2584.
Wang FP, Wang JB, Jian HH, Zhang B, Li SK, Wang F et al. (2008). Environmental adaptation: genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLoS One 3: e1937.
Xiao X, Wang P, Zeng X, Bartlett DH, Wang FP . (2007). Shewanella psychrophila sp. nov. and Shewanella piezotolerans sp. nov., isolated from west pacific deep-sea sediment. Int J Syst Evol Microbiol 57: 60–65.
Acknowledgements
This work has been partly supported by Natural Science Foundation of China (grant number 40625016, 40776095, 40876086), by the National High-Tech Program (2007AA091904), and by COMRA fund (DYXM-115-02-2-03).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, Y., Wang, F., Xu, J. et al. Physiological and evolutionary studies of NAP systems in Shewanella piezotolerans WP3. ISME J 5, 843–855 (2011). https://doi.org/10.1038/ismej.2010.182
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2010.182
Keywords
This article is cited by
-
Transcriptomic Analysis Reveals Common Adaptation Mechanisms Under Different Stresses for Moderately Piezophilic Bacteria
Microbial Ecology (2021)
-
Physiological and transcriptional approaches reveal connection between nitrogen and manganese cycles in Shewanella algae C6G3
Scientific Reports (2017)
-
Characterization of the relationship between polar and lateral flagellar structural genes in the deep-sea bacterium Shewanella piezotolerans WP3
Scientific Reports (2016)
-
Development of a genetic system for the deep-sea psychrophilic bacterium Pseudoalteromonas sp. SM9913
Microbial Cell Factories (2014)
-
Selenite reduction by Shewanella oneidensis MR-1 is mediated by fumarate reductase in periplasm
Scientific Reports (2014)