Abstract
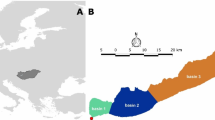
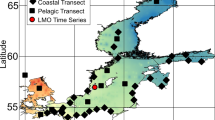
High-throughput sequencing studies during the last decade have uncovered that bacterial genomes are very diverse and dynamic, resulting primarily from the frequent and promiscuous horizontal gene exchange that characterizes the bacterial domain of life. However, a robust understanding of the rates of genetic exchange for most bacterial species under natural conditions and the influence of the ecological settings on the rates remain elusive, severely limiting our view of the microbial world. Here, we analyzed the complete genomic sequences and expressed transcriptomes of several Shewanella baltica isolates recovered from different depths in the Baltic Sea and found that isolates from more similar depths had exchanged a larger fraction of their core and auxiliary genome, up to 20% of the total, compared with isolates from more different depths. The exchanged genes seem to be ecologically important and contribute to the successful adaptation of the isolates to the unique physicochemical conditions of the depth. Importantly, the latter genes were exchanged in very recent past, presumably as an effect of isolate's seasonal migration across the water column, and reflected sexual speciation within the same depth. Therefore, our findings reveal that genetic exchange in response to environmental settings may be surprisingly rapid, which has important broader impacts for understanding bacterial speciation and evolution and for modeling bacterial responses to human-induced environmental impacts.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25: 3389–3402.
Backer H, Leppanen JM, Brusendorff AC, Forsius K, Stankiewicz M, Mehtonen J et al. (2010). HELCOM Baltic Sea action plan—a regional programme of measures for the marine environment based on the Ecosystem approach. Mar Pollut Bull 60: 642–649.
Beiko RG, Harlow TJ, Ragan MA . (2005). Highways of gene sharing in prokaryotes. Proc Natl Acad Sci USA 102: 14332–14337.
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW . (2009). GenBank. Nucleic Acids Res 37: D26–D31.
Brettar I, Moore ER, Hofle MG . (2001). Phylogeny and abundance of novel denitrifying bacteria isolated from the water column of the central Baltic sea. Microb Ecol 42: 295–305.
Bruen TC, Philippe H, Bryant D . (2006). A simple and robust statistical test for detecting the presence of recombination. Genetics 172: 2665–2681.
Caro-Quintero A, Rodriguez-Castano GP, Konstantinidis KT . (2009). Genomic insights into the convergence and pathogenicity factors of Campylobacter jejuni and Campylobacter coli species. J Bacteriol 191: 5824–5831.
Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ et al. (2009). Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA 106: 15442–15447.
Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF et al. (2006). Genomic islands and the ecology and evolution of Prochlorococcus. Science 311: 1768–1770.
Drake JW, Charlesworth B, Charlesworth D, Crow JF . (1998). Rates of spontaneous mutation. Genetics 148: 1667–1686.
Eppley JM, Tyson GW, Getz WM, Banfield JF . (2007). Genetic exchange across a species boundary in the archaeal genus ferroplasma. Genetics 177: 407–416.
Fraser C, Hanage WP, Spratt BG . (2007). Recombination and the nature of bacterial speciation. Science 315: 476–480.
Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS et al. (2008). Towards environmental systems biology of Shewanella. Nat Rev Microbiol 6: 592–603.
Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ et al. (2005). Opinion: re-evaluating prokaryotic species. Nat Rev Microbiol 3: 733–739.
Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245.
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM . (2007). DNA--DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57: 81–91.
Handelsman J, Tiedje J, Alvarez-Cohen L, Ashburner M, Cann I, Delong E et al. (2007). The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet. The National Academies Press: Washington, DC.
Hussain H, Grove J, Griffiths L, Busby S, Cole J . (1994). A seven-gene operon essential for formate-dependent nitrite reduction to ammonia by enteric bacteria. Mol Microbiol 12: 153–163.
Jarvik T, Smillie C, Groisman EA, Ochman H . (2010). Short-term signatures of evolutionary change in the Salmonella enterica serovar typhimurium 14028 genome. J Bacteriol 192: 560–567.
Konstantinidis KT, DeLong EF . (2008). Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J 2: 1052–1065.
Konstantinidis KT, Ramette A, Tiedje JM . (2006a). The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci 361: 1929–1940.
Konstantinidis KT, Ramette A, Tiedje JM . (2006b). Toward a more robust assessment of intraspecies diversity, using fewer genetic markers. Appl Environ Microbiol 72: 7286–7293.
Konstantinidis KT, Serres MH, Romine MF, Rodrigues JL, Auchtung J, McCue LA et al. (2009). Comparative systems biology across an evolutionary gradient within the Shewanella genus. Proc Natl Acad Sci USA 106: 15909–15914.
Konstantinidis KT, Tiedje JM . (2005). Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA 102: 2567–2572.
Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD . (2006a). Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol 23: 1891–1901.
Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD . (2006b). GARD: a genetic algorithm for recombination detection. Bioinformatics 22: 3096–3098.
Lang AS, Beatty JT . (2007). Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol 15: 54–62.
Lawrence JG, Ochman H . (1997). Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol 44: 383–397.
Lawrence JG, Ochman H . (2002). Reconciling the many faces of lateral gene transfer. Trends Microbiol 10: 1–4.
Myers CR, Nealson KH . (1988). Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240: 1319–1321.
Neumann T . (2006). The fate of river-borne nitrogen in the Baltic Sea: an example for the River Oder. Estuar Coast Shelf Sci 73: 1–7.
Retchless AC, Lawrence JG . (2007). Temporal fragmentation of speciation in bacteria. Science 317: 1093–1096.
Sheppard SK, McCarthy ND, Falush D, Maiden MC . (2008). Convergence of Campylobacter species: implications for bacterial evolution. Science 320: 237–239.
Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ‘pan-genome’. Proc Natl Acad Sci USA 102: 13950–13955.
Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43.
Welch RA, Burland V, Plunkett III G, Redford P, Roesch P, Rasko D et al. (2002). Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA 99: 17020–17024.
Wilson DJ, Gabriel E, Leatherbarrow AJ, Cheesbrough J, Gee S, Bolton E et al. (2009). Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol Biol Evol 26: 385–397.
Yang Z . (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591.
Zhaxybayeva O, Gogarten JP, Charlebois RL, Doolittle WF, Papke RT . (2006). Phylogenetic analyses of cyanobacterial genomes: quantification of horizontal gene transfer events. Genome Res 16: 1099–1108.
Ziemke F, Brettar I, Hofle MG . (1997). Stability and diversity of the genetic structure of a Shewanella putrefaciens population in the water column of the central Baltic. Aquat Microb Ecol 13: 63–74.
Acknowledgements
We thank Professors James Tiedje and Frank Loeffler for helpful suggestions regarding the manuscript and the Shewanella Federation for supporting work on Shewanella genomics. Contributions of the Joint Genome Institute for the genome sequences used in this study are also acknowledged. This work is supported by the US Department of Energy under Contract No. DE-FG02-07ER64389.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Caro-Quintero, A., Deng, J., Auchtung, J. et al. Unprecedented levels of horizontal gene transfer among spatially co-occurring Shewanella bacteria from the Baltic Sea. ISME J 5, 131–140 (2011). https://doi.org/10.1038/ismej.2010.93
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2010.93
Keywords
This article is cited by
-
Pan-genome analyses of 24 Shewanella strains re-emphasize the diversification of their functions yet evolutionary dynamics of metal-reducing pathway
Biotechnology for Biofuels (2018)
-
Genome-wide selective sweeps and gene-specific sweeps in natural bacterial populations
The ISME Journal (2016)
-
A Tree of Cellular Life Inferred from a Genomic Census of Molecular Functions
Journal of Molecular Evolution (2014)
-
An automated approach for the identification of horizontal gene transfers from complete genomes reveals the rhizome of Rickettsiales
BMC Evolutionary Biology (2012)
-
Patterns and architecture of genomic islands in marine bacteria
BMC Genomics (2012)