Abstract
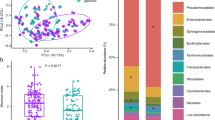
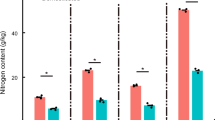
The above- and below-ground parts of rice plants create specific habitats for various microorganisms. In this study, we characterized the phyllosphere and rhizosphere microbiota of rice cultivars using a metaproteogenomic approach to get insight into the physiology of the bacteria and archaea that live in association with rice. The metaproteomic datasets gave rise to a total of about 4600 identified proteins and indicated the presence of one-carbon conversion processes in the rhizosphere as well as in the phyllosphere. Proteins involved in methanogenesis and methanotrophy were found in the rhizosphere, whereas methanol-based methylotrophy linked to the genus Methylobacterium dominated within the protein repertoire of the phyllosphere microbiota. Further, physiological traits of differential importance in phyllosphere versus rhizosphere bacteria included transport processes and stress responses, which were more conspicuous in the phyllosphere samples. In contrast, dinitrogenase reductase was exclusively identified in the rhizosphere, despite the presence of nifH genes also in diverse phyllosphere bacteria.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Anesti V, McDonald IR, Ramaswamy M, Wade WG, Kelly DP, Wood AP . (2005). Isolation and molecular detection of methylotrophic bacteria occurring in the human mouth. Environ Microbiol 7: 1227–1238.
Anthony C, Williams P . (2003). The structure and mechanism of methanol dehydrogenase. BBA - Proteins Proteom 1647: 18–23.
Barea JM, Pozo MJ, Azcon R, Azcon-Aguilar C . (2005). Microbial co-operation in the rhizosphere. J Exp Bot 56: 1761–1778.
Bastida F, Moreno JL, Nicolas C, Hernandez T, Garcia C . (2009). Soil metaproteomics: a review of an emerging environmental science. Significance, methodology and perspectives. Eur J Soil Sci 60: 845–859.
Beattie GA . (2006). Plant-associated bacteria: survey, molecular phylogeny, genomics and recent advances. In: Gnanamanickam SS (ed). Plant-associated bacteria. Springer: Dordrecht, pp 1–56.
Bodelier PL, Roslev P, Henckel T, Frenzel P . (2000). Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature 403: 421–424.
Bosse U, Frenzel P . (1997). Activity and distribution of methane-oxidizing bacteria in flooded rice soil microcosms and in rice plants (Oryza sativa). Appl Environ Microbiol 63: 1199–1207.
Brune A, Frenzel P, Cypionka H . (2000). Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol Rev 24: 691–710.
Chinnadurai C, Balachandar D, Sundaram SP . (2009). Characterization of 1-aminocyclopropane-1-carboxylate deaminase producing methylobacteria from phyllosphere of rice and their role in ethylene regulation. World J Microb Biot 25: 1403–1411.
Conrad R . (2007). Microbial ecology of methanogens and methanotrophs. Adv Agron 96: 1–63.
De Costa DM, Samarasinghe SST, Dias HRD, Dissanayake DMN . (2008). Control of rice sheath blight by phyllosphere epiphytic microbial antagonists. Phytoparasitica 36: 52–65.
Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R et al. (2009). Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci USA 106: 16428–16433.
Derridj S . (1996). Nutrients on the leaf surface. In: Morris CE, Nicot PC, Nguyen-The C (eds). Aerial plant surface microbiology. Plenum Press: New York, pp 25–42.
Elbeltagy A, Ando Y . (2008). Expression of nitrogenase gene (NIFH) in roots and stems of rice, Oryza sativa, by endophytic nitrogen-fixing communities. Afr J Biotechnol 7: 1950–1957.
Elbeltagy A, Nishioka K, Sato T, Suzuki H, Ye B, Hamada T et al. (2001). Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl Environ Microbiol 67: 5285–5293.
Elbeltagy A, Nishioka K, Suzuki H, Sato T, Sato YI, Morisaki H et al. (2000). Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. Soil Sci Plant Nutr 46: 617–629.
Eller G, Frenzel P . (2001). Changes in activity and community structure of methane-oxidizing bacteria over the growth period of rice. Appl Environ Microbiol 67: 2395–2403.
Frenzel P . (2000). Plant-associated methane oxidation in rice fields and wetlands. Adv Microb Ecol 16: 85–114.
Fürnkranz M, Wanek W, Richter A, Abell G, Rasche F, Sessitsch A . (2008). Nitrogen fixation by phyllosphere bacteria associated with higher plants and their colonizing epiphytes of a tropical lowland rainforest of Costa Rica. ISME J 2: 561–570.
Hagemeier CH, Krüer M, Thauer RK, Warkentin E, Ermler U . (2006). Insight into the mechanism of biological methanol activation based on the crystal structure of methanol:cobalamin methyltransferase complex MtaBC from Methanosarcina barkeri. Proc Natl Acad Sci USA 103: 18917–18922.
Hakemian AS, Rosenzweig AC . (2007). The biochemistry of methane oxidation. Annu Rev Biochem 76: 223–241.
Hanson RS, Hanson TE . (1996). Methanotrophic bacteria. Microbiol Rev 60: 439–471.
Hao WY, Ren LX, Ran W, Shen QR . (2010). Allelopathic effects of root exudates from watermelon and rice plants on Fusarium oxysporum f.sp. niveum. Plant Soil 336: 485–497.
Horz HP, Yimga MT, Liesack W . (2001). Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl Environ Microbiol 67: 4177–4185.
Kalyuzhnaya MG, Hristova KR, Lidstrom ME, Chistoserdova L . (2008). Characterization of a novel methanol dehydrogenase in representatives of Burkholderiales: implications for environmental detection of methylotrophy and evidence for convergent evolution. J Bacteriol 190: 3817–3823.
Keppler F, Boros M, Frankenberg C, Lelieveld J, McLeod A, Pirttilä AM et al. (2009). Methane formation in aerobic environments. Environ Chem 6: 459–465.
Kim MC, Ahn JH, Shin HC, Kim T, Ryu TH, Kim DH et al. (2008). Molecular analysis of bacterial community structures in paddy soils for environmental risk assessment with two varieties of genetically modified rice, Iksan 483 and Milyang 204. J Microbiol Biotechnol 18: 207–218.
Knief C, Dunfield PF . (2005). Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environ Microbiol 7: 1307–1317.
Knief C, Delmotte N, Vorholt JA . (2011). Bacterial adaptation to life in assoication with plants–a proteomics perspective from culture to in situ conditions. Proteomics 11: 3086–3105.
Knief C, Vanitchung S, Harvey NW, Conrad R, Dunfield PF, Chidthaisong A . (2005). Diversity of methanotrophic bacteria in tropical upland soils under different land uses. Appl Environ Microbiol 71: 3826–3831.
Kowalchuk GA, Yergeau E, Leveau JHJ, Sessitsch A, Bailey M . (2010). Plant-associated microbial communities. In: Liu W-T, Jansson JK (eds). Environmental molecular microbiology. Caister Academic Press, pp 131–148.
Krüger M, Frenzel P, Conrad R . (2001). Microbial processes influencing methane emission from rice fields. Global Change Biol 7: 49–63.
Ladha JK, Reddy PM . (2000). The quest for nitrogen fixation in rice. International Rice Research Institute: Los Baños, Philippines.
Liesack W, Schnell S, Revsbech NP . (2000). Microbiology of flooded rice paddies. FEMS Microbiol Rev 24: 625–645.
Lu Y, Rosencrantz D, Liesack W, Conrad R . (2006). Structure and activity of bacterial community inhabiting rice roots and the rhizosphere. Environ Microbiol 8: 1351–1360.
Lueders T, Friedrich MW . (2002). Effects of amendment with ferrihydrite and gypsum on the structure and activity of methanogenic populations in rice field soil. Appl Environ Microbiol 68: 2484–2494.
Madhaiyan M, Poonguzhali S, Kwon SW, Sa TM . (2009). Methylobacterium phyllosphaerae sp. nov., a pink-pigmented, facultative methylotroph from the phyllosphere of rice. Int J Syst Evol Microbiol 59: 22–27.
Madhaiyan M, Poonguzhali S, Sa T . (2007). Influence of plant species and environmental conditions on epiphytic and endophytic pink-pigmented facultative methylotrophic bacterial populations associated with field-grown rice cultivars. J Microbiol Biotechnol 17: 1645–1654.
Madhaiyan M, Poonguzhali S, Senthilkumar M, Seshadri S, Chung HY, Yang JC et al. (2004). Growth promotion and induction of systemic resistance in rice cultivar Co-47 (Oryza sativa L.) by Methylobacterium spp. Bot Bull Acad Sin 45: 315–324.
Maliti CM, Basile DV, Corpe WA . (2005). Effects of Methylobacterium spp. strains on rice Oryza sativa L. callus induction, plantlet regeneration, and seedlings growth in vitro. J Torrey Bot Soc 132: 355–367.
Mano H, Tanaka F, Nakamura C, Kaga H, Morisaki H . (2007). Culturable endophytic bacterial flora of the maturing leaves and roots of rice plants (Oryza sativa) cultivated in a paddy field. Microbes Environ 22: 175–185.
Nakatsu CH, Hristova K, Hanada S, Meng XY, Hanson JR, Scow KM et al. (2006). Methylibium petroleiphilum gen. nov., sp. nov., a novel methyl tert-butyl ether-degrading methylotroph of the Betaproteobacteria. Int J Syst Evol Microbiol 56: 983–989.
Nesvizhskii AI, Keller A, Kolker E, Aebersold R . (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658.
Pedraza RO, Bellone CH, de Bellone S, Sorte PMB, Teixeira KRD . (2009). Azospirillum inoculation and nitrogen fertilization effect on grain yield and on the diversity of endophytic bacteria in the phyllosphere of rice rainfed crop. Eur J Soil Biol 45: 36–43.
Prakamhang J, Minamisawa K, Teamtaisong K, Boonkerd N, Teaumroong N . (2009). The communities of endophytic diazotrophic bacteria in cultivated rice (Oryza sativa L.). Appl Soil Ecol 42: 141–149.
Prasanna R, Nain L, Pandey AK, Nayak S . (2010). Exploring the ecological significance of microbial diversity and networking in the rice ecosystem. In: Dion P (ed). Soil biology and agriculture in the tropics. Springer Berlin Heidelberg, pp 139–161.
Schmidt S, Christen P, Kiefer P, Vorholt JA . (2010). Functional investigation of methanol dehydrogenase-like protein XoxF in Methylobacterium extorquens AM1. Microbiology 156: 2575–2586.
Shrestha M, Abraham WR, Shrestha PM, Noll M, Conrad R . (2008). Activity and composition of methanotrophic bacterial communities in planted rice soil studied by flux measurements, analyses of pmoA gene and stable isotope probing of phospholipid fatty acids. Environ Microbiol 10: 400–412.
Shrestha PM, Kube M, Reinhardt R, Liesack W . (2009). Transcriptional activity of paddy soil bacterial communities. Environ Microbiol 11: 960–970.
Stark M, Berger SA, Stamatakis A, von Mering C . (2010). MLTreeMap - accurate Maximum Likelihood placement of environmental DNA sequences into taxonomic and functional reference phylogenies. BMC Genomics 11: 461.
Suzuki R, Shimodaira H . (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542.
Sy A, Timmers AC, Knief C, Vorholt JA . (2005). Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl Environ Microbiol 71: 7245–7252.
Thauer RK . (1998). Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144: 2377–2406.
von Ophem PW, van Beeumen J, Duine JA . (1993). Nicotinoprotein [NAD(P)-containing] alcohol/aldehyde oxidoreductases. Purification and characterization of a novel type from Amycolatopsis methanolica. Eur J Biochem 212: 819–826.
Vorholt JA, Marx CJ, Lidstrom ME, Thauer RK . (2000). Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J Bacteriol 182: 6645–6650.
Wang HB, Zhang ZX, Li H, He HB, Fang CX, Zhang AJ et al. (2011). Characterization of metaproteomics in crop rhizospheric soil. J Proteome Res 10: 932–940.
Wassmann R, Aulakh MS . (2000). The role of rice plants in regulating mechanisms of methane missions. Biol Fertil Soils 31: 20–29.
Watanabe I, Hashimoto T, Shimoyama A . (1997). Methane-oxidizing activities and methanotrophic populations associated with wetland rice plants. Biol Fertil Soils 24: 261–265.
Wu LK, Wang HB, Zhang ZX, Lin R, Zhang ZY, Lin WX . (2011). Comparative metaproteomic analysis on consecutively Rehmannia glutinosa-monocultured rhizosphere soil. Plos One 6: e20611.
Wu LQ, Ma K, Lu YH . (2009). Rice roots select for type I methanotrophs in rice field soil. Syst Appl Microbiol 32: 421–428.
Yang JH, Liu HX, Zhu GM, Pan YL, Xu LP, Guo JH . (2008). Diversity analysis of antagonists from rice-associated bacteria and their application in biocontrol of rice diseases. J Appl Microbiol 104: 91–104.
Zhu W, Reich CI, Olsen GJ, Giometti CS, Yates 3rd JR . (2004). Shotgun proteomics of Methanococcus jannaschii and insights into methanogenesis. J Proteome Res 3: 538–548.
Acknowledgements
We thank Agnes Padre and Enrique Montserrat for support at the International Rice Research Institute during sample collection. We thank FGCZ for access to the proteomics facility and support. The study was supported by ETH Zurich.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Knief, C., Delmotte, N., Chaffron, S. et al. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J 6, 1378–1390 (2012). https://doi.org/10.1038/ismej.2011.192
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2011.192
Keywords
This article is cited by
-
Characterization of phyllosphere endophytic lactic acid bacteria reveals a potential novel route to enhance silage fermentation quality
Communications Biology (2024)
-
Rhizosphere Microorganisms and Soil Physicochemical Properties of Restored Wetland Plant Communities at Cutting Slash of Populus deltoides in Dongting Lake
Wetlands (2023)
-
Contribution of Leaf-Associated Microorganisms from Native Andean Ericaceae against Botrytis cinerea in Vaccinium corymbosum Cultivars
Journal of Soil Science and Plant Nutrition (2023)
-
Nitrogen cycling-related functional genes exhibit higher sensibility in soil than leaf phyllosphere of different tree species in the subtropical forests
Plant and Soil (2023)