Abstract
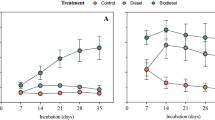
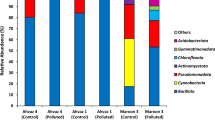
Increased exploration and exploitation of resources in the Arctic is leading to a higher risk of petroleum contamination. A number of Arctic microorganisms can use petroleum for growth-supporting carbon and energy, but traditional approaches for stimulating these microorganisms (for example, nutrient addition) have varied in effectiveness between sites. Consistent environmental controls on microbial community response to disturbance from petroleum contaminants and nutrient amendments across Arctic soils have not been identified, nor is it known whether specific taxa are universally associated with efficient bioremediation. In this study, we contaminated 18 Arctic soils with diesel and treated subsamples of each with monoammonium phosphate (MAP), which has successfully stimulated degradation in some contaminated Arctic soils. Bacterial community composition of uncontaminated, diesel-contaminated and diesel+MAP soils was assessed through multiplexed 16S (ribosomal RNA) rRNA gene sequencing on an Ion Torrent Personal Genome Machine, while hydrocarbon degradation was measured by gas chromatography analysis. Diversity of 16S rRNA gene sequences was reduced by diesel, and more so by the combination of diesel and MAP. Actinobacteria dominated uncontaminated soils with <10% organic matter, while Proteobacteria dominated higher-organic matter soils, and this pattern was exaggerated following disturbance. Degradation with and without MAP was predictable by initial bacterial diversity and the abundance of specific assemblages of Betaproteobacteria, respectively. High Betaproteobacteria abundance was positively correlated with high diesel degradation in MAP-treated soils, suggesting this may be an important group to stimulate. The predictability with which bacterial communities respond to these disturbances suggests that costly and time-consuming contaminated site assessments may not be necessary in the future.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Aislabie J, Saul DJ, Foght JM . (2006). Bioremediation of hydrocarbon-contaminated polar soils. Extremophiles 10: 171–179.
Allison SD, Hanson CA, Treseder KK . (2007). Nitrogen fertilization reduces diversity and alters community structure of active fungi in boreal ecosystems. Soil Biol Biochem 39: 1878–1887.
Atlas RM . (1986). Fate of petroleum pollutants in Arctic ecosystems. Water Sci Technol 18: 59–67.
Baker GC, Smith JJ, Cowan DA . (2003). Review and re-analysis of domain-specific 16S primers. J Microbiol Meth 55: 541–555.
Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK . (2005). The contribution of species richness and composition to bacterial services. Nature 436: 1157–1160.
Bell TH, Yergeau E, Martineau C, Juck D, Whyte LG, Greer CW . (2011). Identification of nitrogen-incorporating bacteria in petroleum-contaminated Arctic soils by using [15N]DNA-based stable isotope probing and pyrosequencing. Appl Environ Microbiol 77: 4163–4171.
Björk RG, Björkman MP, Andersson MX, Klemedtsson L . (2008). Temporal variation in soil microbial communities in Alpine tundra. Soil Biol Biochem 40: 266–268.
Caro-Quintero A, Konstantinidis KT . (2012). Bacterial species may exist, metagenomics reveal. Environ Microbiol 14: 347–355.
Castorena G, Mugica V, Le Borgne S, Acuña ME, Bustos-Jaimes I, Aburto J . (2006). Carbazole biodegradation in gas oil/water biphasic media by a new isolated bacterium Burkholderia sp strain IMP5GC. J Appl Microbiol 100: 739–745.
Chang WJ, Klemm S, Beaulieu C, Hawari J, Whyte L, Ghoshal S . (2011). Petroleum hydrocarbon biodegradation under seasonal freeze-thaw soil temperature regimes in contaminated soils from a sub-Arctic site. Environ Sci Technol 45: 1061–1066.
Chu HY, Fierer N, Lauber CL, Caporaso JG, Knight R, Grogan P . (2010). Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol 12: 2998–3006.
Claesson MJ, O'Sullivan O, Wang Q, Nikkila J, Marchesi JR, Smidt H et al. (2009). Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4: e6669.
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R . (2009). Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697.
Dorn PB, Salanitro JP . (2000). Temporal ecological assessment of oil contaminated soils before and after bioremediation. Chemosphere 40: 419–426.
Fargione J, Tilman D, Dybzinski R, Lambers JH, Clark C, Harpole WS et al. (2007). From selection to complementarity: shifts in the causes of biodiversity-productivity relationships in a long-term biodiversity experiment. Proc Roy Soc B-Biol Sci 274: 871–876.
Fierer N, Schimel JP, Holden PA . (2003). Variations in microbial community composition through two soil depth profiles. Soil Biol Biochem 35: 167–176.
Fierer N, Jackson RB . (2006). The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103: 626–631.
Fierer N, Bradford MA, Jackson RB . (2007). Toward an ecological classification of soil bacteria. Ecology 88: 1354–1364.
Fortunato CS, Herfort L, Zuber P, Baptista AM, Crump BC . (2012). Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. ISME J 6: 554–563.
Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S . (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA 103: 13104–13109.
Greer CW . (2009). Bioremediation of contaminated sites in the Canadian Arctic: monitoring performance and the effects of biostimulation using molecular methods. In: Bej AK, Aislabie J, Atlas RM, (eds) Polar Microbiology: The Ecology, Diversity, And Bioremediation Potential of Microorganisms in Extremely Cold Environments. CRC Press, Inc.: Boca Raton, FL, pp 319–338.
Greer CW, Whyte LG, Niederberger TD . (2010). Microbial communities in hydrocarbon-contaminated temperate, tropical, alpine, and polar soils. In: Timmis KN, (eds) Handbook of Hydrocarbon and Lipid Microbiology. Springer: Berlin Heidelberg, pp 2313–2328.
Griffiths BS, Ritz K, Ebblewhite N, Dobson G . (1999). Soil microbial community structure: effects of substrate loading rates. Soil Biol Biochem 31: 145–153.
Haddad NM, Holyoak M, Mata TM, Davies KF, Melbourne BA, Preston K . (2008). Species' traits predict the effects of disturbance and productivity on diversity. Ecol Lett 11: 348–356.
Heczko U, Abe A, Finlay BB . (2000). Segmented filamentous bacteria prevent colonization of enteropathogenic Escherichia coli O103 in rabbits. J Infect Dis 181: 1027–1033.
Hemme CL, Deng Y, Gentry TJ, Fields MW, Wu LY, Barua S et al. (2010). Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J 4: 660–672.
Hesselsoe M, Bjerring ML, Henriksen K, Loll P, Nielsen JL . (2008). Method for measuring substrate preferences by individual members of microbial consortia proposed for bioaugmentation. Biodegradation 19: 621–633.
Horner-Devine MC, Carney KM, BJM Bohannan . (2004). An ecological perspective on bacterial biodiversity. Proc Roy Soc Lond B Bio 271: 113–122.
Kang YS, Park W . (2010). Protection against diesel oil toxicity by sodium chloride-induced exopolysaccharides in Acinetobacter sp strain DR1. J Biosci Bioeng 109: 118–123.
Keiser AD, Strickland MS, Fierer N, Bradford MA . (2011). The effect of resource history on the functioning of soil microbial communities is maintained across time. Biogeosciences 8: 1477–1486.
Lauber CL, Hamady M, Knight R, Fierer N . (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75: 5111–5120.
Lennon JT, Cottingham KL . (2008). Microbial productivity in variable resource environments. Ecology 89: 1001–1014.
Ma YF, Wang L, Shao ZZ . (2006). Pseudomonas, the dominant polycyclic aromatic hydrocarbon-degrading bacteria isolated from Antarctic soils and the role of large plasmids in horizontal gene transfer. Environ Microbiol 8: 455–465.
Magurran AE, Henderson PA . (2010). Temporal turnover and the maintenance of diversity in ecological assemblages. Philos T R Soc B 365: 3611–3620.
Mbadinga SM, Wang LY, Zhou L, Liu JF, Gu JD, Mu BZ . (2011). Microbial communities involved in anaerobic degradation of alkanes. Int Biodeter Biodegr 65: 1–13.
McCarthy K, Walker L, Vigoren L, Bartel J . (2004). Remediation of spilled petroleum hydrocarbons by in situ landfarming at an arctic site. Cold Reg Sci Technol 40: 31–39.
Neff JM, Ostazeski S, Gardiner W, Stejskal I . (2000). Effects of weathering on the toxicity of three offshore Australian crude oils and a diesel fuel to marine animals. Environ Toxicol Chem 19: 1809–1821.
Owsianiak M, Szulc A, Chrzanowski L, Cyplik P, Bogacki M, Olejnik-Schmidt AK et al. (2009). Biodegradation and surfactant-mediated biodegradation of diesel fuel by 218 microbial consortia are not correlated to cell surface hydrophobicity. Appl Microbiol Biot 84: 545–553.
Pepi M, Minacci A, Di Cello F, Baldi F, Fani R . (2003). Long-term analysis of diesel fuel consumption in a co-culture of Acinetobacter venetianus, Pseudomonas putida and Alcaligenes faecalis. Anton Leeuw Int J G 83: 3–9.
Peter H, Beier S, Bertilsson S, Lindström ES, Langenheder S, Tranvik LJ . (2011). Function-specific response to depletion of microbial diversity. ISME J 5: 351–361.
Pommier T, Douzery EJP, Mouillot D . (2012). Environment drives high phylogenetic turnover among oceanic bacterial communities. Biol Lett 8: 562–566.
Powell SM, Bowman JP, Ferguson SH, Snape I . (2010). The importance of soil characteristics to the structure of alkane-degrading bacterial communities on sub-Antarctic Macquarie Island. Soil Biol Biochem 42: 2012–2021.
Ramirez KS, Craine JM, Fierer N . (2012). Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Change Biol 18: 1918–1927.
Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-González MI, Rojas A et al. (2002). Mechanisms of solvent tolerance in Gram-negative bacteria. Annu Rev Microbiol 56: 743–768.
Salles JF, Poly F, Schmid B, Le Roux X . (2009). Community niche predicts the functioning of denitrifying bacterial assemblages. Ecology 90: 3324–3332.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb 75: 7537–7541.
Schulz S, Giebler J, Chatzinotas A, Wick LY, Fetzer I, Welzl G et al. (2012). Plant litter and soil type drive abundance, activity and community structure of alkB harbouring microbes in different soil compartments. ISME J 6: 1763–1774.
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Thomassin-Lacroix EJM, Eriksson M, Reimer KJ, Mohn WW . (2002). Biostimulation and bioaugmentation for on-site treatment of weathered diesel fuel in Arctic soil. Appl Microbiol Biot 59: 551–556.
Thompson IP, van der Gast CJ, Ciric L, Singer AC . (2005). Bioaugmentation for bioremediation: the challenge of strain selection. Environ Microbiol 7: 909–915.
van Herwijnen R, Joffe B, Ryngaert A, Hausner M, Springael D, Govers HAJ et al. (2006). Effect of bioaugmentation and supplementary carbon sources on degradation of polycyclic aromatic hydrocarbons by a soil-derived culture. FEMS Microbiol Ecol 55: 122–135.
Violle C, Pu ZC, Jiang L . (2010). Experimental demonstration of the importance of competition under disturbance. Proc Natl Acad Sci USA 107: 12925–12929.
Walworth J, Braddock J, Woolard C . (2001). Nutrient and temperature interactions in bioremediation of cryic soils. Cold Reg Sci Technol 32: 85–91.
Walworth JL, Woolard CR, Braddock JF, Reynolds CM . (1997). Enhancement and inhibition of soil petroleum biodegradation through the use of fertilizer nitrogen: an approach to determining optimum levels. J Soil Contam 6: 465–480.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267.
Whyte LG, Bourbonnière L, Greer CW . (1997). Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways. Appl Environ Microbiol 63: 3719–3723.
Yergeau E, Bokhorst S, Huiskes AHL, Boschker HTS, Aerts R, Kowalchuk GA . (2007). Size and structure of bacterial, fungal and nematode communities along an Antarctic environmental gradient. FEMS Microbiol Ecol 59: 436–451.
Yergeau E, Arbour M, Brousseau R, Juck D, Lawrence JR, Masson L et al. (2009). Microarray and real-time PCR analyses of the responses of high-Arctic soil bacteria to hydrocarbon pollution and bioremediation treatments. Appl Environ Microbiol 75: 6258–6267.
Yergeau E, Bokhorst S, Kang S, Zhou JZ, Greer CW, Aerts R et al. (2012a). Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. ISME J 6: 692–702.
Yergeau E, Lawrence JR, Sanschagrin S, Waiser MJ, Korber DR, Greer CW . (2012b). Next-generation sequencing of microbial communities in the Athabasca river and its tributaries in relation to oil sands mining activities. Appl Environ Microbiol 78: 7626–7637.
Yergeau E, Sanschagrin S, Beaumier D, Greer CW . (2012c). Metagenomic analysis of the bioremediation of diesel-contaminated Canadian high Arctic soils. PLoS One 7: e30058.
Acknowledgements
We thank Matt Wallenstein, Claudia Boot, Nicolas Lecomte, Emilie Champagne, Jen Allan, Isabelle Laurion, Chantal Lemieux, Karita Neghandi, Philippe Galipeau, Marie-Jeanne Rioux, Katryne Larrivée, JF Lamarre, Elise Bolduc, and the Center d’Études Nordiques for soil collection, Julie Champagne for sequencing assistance, and Chantale Beaulieu and Stéphane Deschamps for assistance with GC analysis. This work was supported by the NRCan PERD Program, and an NSERC postgraduate scholarship to TH Bell.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Bell, T., Yergeau, E., Maynard, C. et al. Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J 7, 1200–1210 (2013). https://doi.org/10.1038/ismej.2013.1
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2013.1
Keywords
This article is cited by
-
Microbial remediation of petroleum-contaminated soil focused on the mechanism and microbial response: a review
Environmental Science and Pollution Research (2024)
-
Autochthonous psychrophilic hydrocarbonoclastic bacteria and its ecological function in contaminated cold environments
Biodegradation (2024)
-
Prokaryotic diversity and community composition in the surface sediments of the Changjiang River Estuary in summer
Acta Oceanologica Sinica (2023)
-
A comprehensive study on diesel oil bioremediation under microcosm conditions using a combined microbiological, enzymatic, mass spectrometry, and metabarcoding approach
Environmental Science and Pollution Research (2023)
-
Response of soil microecology to different cropping practice under Bupleurum chinense cultivation
BMC Microbiology (2022)