Abstract
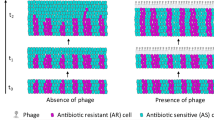
The long-term persistence of antibiotic-resistant bacteria depends on their fitness relative to other genotypes in the absence of drugs. Outside the laboratory, viruses that parasitize bacteria (phages) are ubiquitous, but costs of antibiotic resistance are typically studied in phage-free experimental conditions. We used a mathematical model and experiments with Escherichia coli to show that lytic phages strongly affect the incidence of antibiotic resistance in drug-free conditions. Under phage parasitism, the likelihood that antibiotic-resistant genetic backgrounds spread depends on their initial frequency, mutation rate and intrinsic growth rate relative to drug-susceptible genotypes, because these parameters determine relative rates of phage-resistance evolution on different genetic backgrounds. Moreover, the average cost of antibiotic resistance in terms of intrinsic growth in the antibiotic-free experimental environment was small relative to the benefits of an increased mutation rate in the presence of phages. This is consistent with our theoretical work indicating that, under phage selection, typical costs of antibiotic resistance can be outweighed by realistic increases in mutability if drug resistance and hypermutability are genetically linked, as is frequently observed in clinical isolates. This suggests the long-term distribution of antibiotic resistance depends on the relative rates at which different lineages adapt to other types of selection, which in the case of phage parasitism is probably extremely common, as well as costs of resistance inferred by classical in vitro methods.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Abedon ST, Culler RR . (2007). Bacteriophage evolution given spatial constraint. J Theor Biol 248: 111–119.
Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM . (2011). Phage treatment of human infections. Bacteriophage 1: 66–85.
Alexander HK, Bonhoeffer S . (2012). Pre-existence and emergence of drug resistance in a generalized model of intra-host viral dynamics. Epidemics 4: 187–202.
Andersson DI, Levin BR . (1999). The biological cost of antibiotic resistance. Curr Opin Microbiol 2: 489–493.
Andersson DI, Hughes D . (2010). Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8: 260–271.
Andersson DI, Hughes D . (2011). Persistence of antibiotic resistance in bacterial populations. FEMS Microbiol Rev 35: 901–911.
Angst D, Hall AR . (2013). The cost of antibiotic resistance depends on evolutionary history in Escherichia coli. BMC Evol Biol 13: 163.
Baquero F, Negri MC . (1997). Selective compartments for resistant microorganisms in antibiotic gradients. Bioessays 19: 731–736.
Baquero MR, Galán JC, Turrientes MD, Cantón R, Coque TM, Martínez JL et al. (2005). Increased mutation frequencies in Escherichia coli isolates harboring extended-spectrum β-lactamases. Antimicrob Agents Chemother 49: 4754–4756.
Barton NH . (1995). Linkage and the limits to natural selection. Genetics 140: 821–841.
Barton NH . (2000). Genetic hitchhiking. Philos Trans R Soc Lond B Biol Sci 355: 1553–1562.
Bergstrom CT, Feldgarden M . (2008). The ecology and evolution of antibiotic-resistant bacteria. In: Stearns SC, Koella JC (eds) Evolution in Health and Disease 2nd edn. Oxford University Press: Oxford, pp 125–137.
Björkman J, Andersson DI . (2000). The cost of antibiotic resistance from a bacterial perspective. Drug Resist Updat 3: 237–245.
Björkman J, Nagaev I, Berg OG, Hughes D, Andersson DI . (2000). Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287: 1479–1482.
Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P et al. (2003). Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 185: 6220–6223.
Brockhurst MA, Morgan AD, Fenton A, Buckling A . (2007). Experimental coevolution with bacteria and phage. The Pseudomonas fluorescens—Φ2 model system. Infect Genet Evol 7: 547–552.
Brüssow H, Hendrix RW . (2002). Phage genomics: small is beautiful. Cell 108: 13–16.
Brüssow H . (2005). Phage therapy: the Escherichia coli experience. Microbiology 151: 2133–2140.
Buckling A, Wei Y, Massey RC, Brockhurst MA, Hochberg ME . (2006). Antagonistic coevolution with parasites increases the cost of host deleterious mutations. Proc R Soc B Biol Sci 273: 45–49.
Chanishvili N, Chanishvili T, Tediashvili M, Barrow PA . (2001). Phages and their application against drug-resistant bacteria. J Chem Technol Biot 76: 689–699.
Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brüssow H . (2004). Phage-host interaction: an ecological perspective. J Bacteriol 186: 3677–3686.
Chopra I, O'Neill AJ, Miller K . (2003). The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist Updat 6: 137–145.
Constantinou A, Voelkel-Meiman K, Sternglanz R, McCorquodale MM, McCorquodale DJ . (1986). Involvement of host DNA gyrase in growth of bacteriophage T5. J Virol 57: 875–882.
Cooper TF, Lenski RE, Elena SF . (2005). Parasites and mutational load: an experimental test of a pluralistic theory for the evolution of sex. Proc R Soc B Biol Sci 272: 311–317.
D'Costa VM, McGrann KM, Hughes DW, Wright GD . (2006). Sampling the antibiotic resistome. Science 311: 374–377.
Denamur E, Bonacorsi S, Giraud A, Duriez P, Hilali F, Amorin C et al. (2002). High frequency of mutator strains among human uropathogenic Escherichia coli isolates. J Bacteriol 184: 605–609.
Escobar-Páramo P, Gougat-Barbera C, Hochberg ME . (2012). Evolutionary dynamics of separate and combined exposure of Pseudomonas fluorescens SBW25 to antibiotics and bacteriophage. Evol Appl 5: 583–592.
Fothergill JL, Mowat E, Ledson MJ, Walshaw MJ, Winstanley C . (2010). Fluctuations in phenotypes and genotypes within populations of Pseudomonas aeruginosa in the cystic fibrosis lung during pulmonary exacerbations. J Med Microbiol 59: 472–481.
Foweraker JE, Laughton CR, Brown DF, Bilton D . (2005). Phenotypic variability of Pseudomonas aeruginosa in sputa from patients with acute infective exacerbation of cystic fibrosis and its impact on the validity of antimicrobial susceptibility testing. J Antimicrob Chemother 55: 921–927.
Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJM . (2006). The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312: 1944–1946.
Giraud A, Matic I, Radman M, Fons M, Taddei F . (2002). Mutator bacteria as a risk factor in treatment of infectious diseases. Antimicrob Agents Chemother 46: 863–865.
Gómez P, Buckling A . (2013). Coevolution with phages does not influence the evolution of bacterial mutation rates in soil. ISME J 7: 2242–2244.
Haag CR, Sakwinska O, Ebert D . (2003). Test of synergistic interaction between infection and inbreeding in Daphnia magna. Evolution 57: 777–783.
Hall AR . (2013). Genotype-by-environment interactions due to adaptation and antibiotic resistance in Escherichia coli. J Evol Biol 26: 1655–1664.
Hammer K, Jensen KF, Poulsen P, Oppenheim AB, Gottesman M . (1987). Isolation of Escherichia coli rpoB mutants resistant to killing by lambda cII protein and altered in pyrE gene attenuation. J Bacteriol 169: 5289–5297.
Hancock RE, Reeves P . (1975). Bacteriophage resistance in Escherichia coli K-12: general pattern of resistance. J Bacteriol 121: 983–993.
Hennes KP, Simon M . (1995). Significance of bacteriophages for controlling bacterioplankton growth in a mesotrophic lake. Appl Environ Microbiol 61: 333–340.
Howard RS, Lively CM . (1994). Parasitism, mutation accumulation and the maintenance of sex. Nature 367: 554–557.
Jalasvuori M, Friman VP, Nieminen A, Bamford JKH, Buckling A . (2011). Bacteriophage selection against a plasmid-encoded sex apparatus leads to the loss of antibiotic-resistance plasmids. Biol Lett 7: 902–905.
Johnson T, Barton NH . (2002). The effect of deleterious alleles on adaptation in asexual populations. Genetics 162: 395–411.
Jolivet-Gougeon A, Kovacs B, Le Gall-David S, Le Bars H, Bousarghin L, Bonnaure-Mallet M et al. (2011). Bacterial hypermutation: clinical implications. J Med Microbiol 60: 563–573.
Kaper JB, Nataro JP, Mobley HL . (2004). Pathogenic Escherichia coli. Nat Rev Microbiol 2: 123–140.
Koskella B . (2013). Phage-mediated selection on microbiota of a long-lived host. Curr Biol 23: 1256–1260.
Labrie SJ, Samson JE, Moineau S . (2010). Bacteriophage resistance mechanisms. Nat Rev Microbiol 8: 317–327.
LeClerc JE, Li BG, Payne WL, Cebula TA . (1996). High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274: 1208–1211.
Lenski RE, Rose MR, Simpson SC, Tadler SC . (1991). Long-term experimental evolution in Escherichia coli 1. Adaptation and divergence during 2,000 generations. Am Nat 138: 1315–1341.
Levin BR, Bull JJ . (2004). Population and evolutionary dynamics of phage therapy. Nat Rev Microbiol 2: 166–173.
Levin BR, Moineau S, Bushman M, Barrangou R . (2013). The population and evolutionary dynamics of phage and bacteria with CRISPR-mediated immunity. PLoS Genet 9: e1003312.
Levy SB, Marshall B . (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10: S122–S129.
Li B, Tsui HC, LeClerc JE, Dey M, Winkler ME, Cebula TA . (2003). Molecular analysis of mutS expression and mutation in natural isolates of pathogenic Escherichia coli. Microbiology 149: 1323–1331.
Lieberman MM, Markovitz A . (1970). Depression of guanosine diphosphate-mannose pyrophosphorylase by mutations in two different regulator genes involved in capsular polysaccharide synthesis in Escherichia coli K-12. J Bacteriol 101: 965–972.
Lopez-Pascua LDC, Buckling A . (2008). Increasing productivity accelerates host-parasite coevolution. J Evol Biol 21: 853–860.
Maciá MD, Blanquer D, Togores B, Sauleda J, Pérez JL, Oliver A . (2005). Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob Agents Chemother 49: 3382–3386.
Martin G, Aguilee R, Ramsayer J, Kaltz O, Ronce O . (2013). The probability of evolutionary rescue: towards a quantitative comparison between theory and evolution experiments. Phil Trans R Soc B 368: 20120088.
Martinez JL . (2009). The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc R Soc B Biol Sci 276: 2521–2530.
Maynard Smith J, Haigh J . (1974). The hitch-hiking effect of a favourable gene. Genet Res 23: 23–35.
Mizoguchi K, Morita M, Fischer CR, Yoichi M, Tanji Y, Unno H . (2003). Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Appl Environ Microbiol 69: 170–176.
Morgan AD, Quigley BJ, Brown SP, Buckling A . (2012). Selection on non-social traits limits the invasion of social cheats. Ecol Lett 15: 841–846.
O'Brien S, Rodrigues AM, Buckling A . (2013). The evolution of bacterial mutation rates under simultaneous selection by interspecific and social parasitism. Proc Biol Sci 280: 20131913.
Oliver A, Canton R, Campo P, Baquero F, Blázquez J . (2000). High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288: 1251–1253.
Oliver A, Baquero F, Blázquez J . (2002). The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol Microbiol 43: 1641–1650.
Oliver A, Levin BR, Juan C, Baquero F, Blazquez J . (2004). Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob Agents Chemother 48: 4226–4233.
Pál C, Maciá MD, Oliver A, Schachar I, Buckling A . (2007). Coevolution with viruses drives the evolution of bacterial mutation rates. Nature 450: 1079–1081.
Parzen E . (1999) Stochastic Processes. Society for Industrial and Applied Mathematics: Philadelphia, PA.
Peck JR . (1994). A ruby in the rubbish: beneficial mutations, deleterious mutations and the evolution of sex. Genetics 137: 597–606.
R Development Core Team. (2010) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
Reynolds MG . (2000). Compensatory evolution in rifampin-resistant Escherichia coli. Genetics 156: 1471–1481.
Sander P, Springer B, Prammananan T, Sturmfels A, Kappler M, Pletschette M et al. (2002). Fitness cost of chromosomal drug resistance-conferring mutations. Antimicrob Agents Chemother 46: 1204–1211.
Sapelli RV, Goebel WF . (1964). The capsular polysaccharide of a mucoid variant of E. Coli K 12. Proc Natl Acad Sci USA 52: 265–271.
Scanlan PD, Buckling A . (2012). Co-evolution with lytic phage selects for the mucoid phenotype of Pseudomonas fluorescens SBW25. ISME J 6: 1148–1158.
Scanvic-Hameg A, Chachaty E, Rey J, Pousson C, Ozoux ML, Brunel E et al. (2002). Impact of quinupristin/dalfopristin (RP59500) on the faecal microflora in healthy volunteers. J Antimicrob Chemother 49: 135–139.
Schmitt CK, Kemp P, Molineux IJ . (1995). Streptomycin- and rifampin-resistant mutants of Escherichia coli perturb F-exclusion of bacteriophage-T7 by affecting synthesis of the F-plasmid protein PifA. J Bacteriol 177: 1589–1594.
Seppälä H, Klaukka T, Vuopio-Varkila J, Muotiala A, Helenius H, Lager K et al. (1997). The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. Finnish Study Group for Antimicrobial Resistance. N Engl J Med 337: 441–446.
Sutherland IW, Hughes KA, Skillman LC, Tait K . (2004). The interaction of phage and biofilms. FEMS Microbiol Lett 232: 1–6.
Tazzyman SJ, Bonhoeffer S . (2014). Plasmids and evolutionary rescue by drug resistance. Evolution 68: 2066–2078.
Tenaillon O, Skurnik D, Picard B, Denamur E . (2010). The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8: 207–217.
Thingstad TF, Lignell R . (1997). Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat Microb Ecol 13: 19–27.
Travisano M, Lenski RE . (1996). Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics 143: 15–26.
Trindade S, Sousa A, Xavier KB, Dionisio F, Ferreira MG, Gordo I . (2009). Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet 5: e1000578.
Vos M, Birkett PJ, Birch E, Griffiths RI, Buckling A . (2009). Local adaptation of bacteriophages to their bacterial hosts in soil. Science 325: 833–833.
Waite AJ, Shou W . (2012). Adaptation to a new environment allows cooperators to purge cheaters stochastically. Proc Natl Acad Sci USA 109: 19079–19086.
Webb JS, Lau M, Kjelleberg S . (2004). Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol 186: 8066–8073.
Weinbauer MG . (2004). Ecology of prokaryotic viruses. Fems Microbiol Rev 28: 127–181.
Weinbauer MG, Rassoulzadegan F . (2004). Are viruses driving microbial diversification and diversity? Environ Microbiol 6: 1–11.
West SA, Lively CM, Read AF . (1999). A pluralist approach to sex and recombination. J Evol Biol 12: 1003–1012.
Workentine ML, Sibley CD, Glezerson B, Purighalla S, Norgaard-Gron JC, Parkins MD et al. (2013). Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLoS One 8: e60225.
Zhang QG, Buckling A . (2012). Phages limit the evolution of bacterial antibiotic resistance in experimental microcosms. Evol Appl 5: 575–582.
Zhang QG . (2014). Exposure to phages has little impact on the evolution of bacterial antibiotic resistance on drug concentration gradients. Evol Appl 7: 394–402.
Acknowledgements
Martin Ackermann, Helen Alexander, Sebastian Bonhoeffer, Angus Buckling, Louis Du Plessis, Pedro Gómez-López and Rolf Kümmerli provided helpful comments and discussion. We thank Arnaud Gutierrez and Ivan Matic for the Δara marker, Dominik Refardt for phages and Tobias Bergmiller for the mutator. Sequencing was done at the Genetic Diversity Centre, ETH Zürich. ARH was funded by a Marie Curie Intra-European Fellowship and the Swiss National Science Foundation and SJT by the European Research Council under the 7th Framework Programme of the European Commission (PBDR: Grant Agreement Number 268540).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Tazzyman, S., Hall, A. Lytic phages obscure the cost of antibiotic resistance in Escherichia coli. ISME J 9, 809–820 (2015). https://doi.org/10.1038/ismej.2014.176
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2014.176
This article is cited by
-
Mutant fixation in the presence of a natural enemy
Nature Communications (2023)
-
Phage–Antibiotic Synergy Inhibited by Temperate and Chronic Virus Competition
Bulletin of Mathematical Biology (2022)