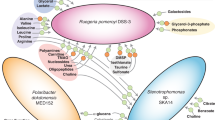
Abstract
Phenotypic plasticity (PP) is the development of alternate phenotypes of a given taxon as an adaptation to environmental conditions. Methodological limitations have restricted the quantification of PP to the measurement of a few traits in single organisms. We used metatranscriptomic libraries to overcome these challenges and estimate PP using the expressed genes of multiple heterotrophic organisms as a proxy for traits in a microbial community. The metatranscriptomes captured the expression response of natural marine bacterial communities grown on differing carbon resource regimes in continuous cultures. We found that taxa with different magnitudes of PP coexisted in the same cultures, and that members of the order Rhodobacterales had the highest levels of PP. In agreement with previous studies, our results suggest that continuous culturing may have specifically selected for taxa featuring a rather high range of PP. On average, PP and abundance changes within a taxon contributed equally to the organism’s change in functional gene abundance, implying that both PP and abundance mediated observed differences in community function. However, not all functional changes due to PP were directly reflected in the bulk community functional response: gene expression changes in individual taxa due to PP were partly masked by counterbalanced expression of the same gene in other taxa. This observation demonstrates that PP had a stabilizing effect on a community’s functional response to environmental change.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Auld JR, Agrawal AA, Relyea RA . (2010). Re-evaluating the costs and limits of adaptive phenotypic plasticity. Proc R Soc B Biol Sci 277: 503–511.
Beier S, Rivers AR, Moran MA, Obernosterer I . (2014). The transcriptional response of prokaryotes to phytoplankton-derived DOM in seawater. Environ Microbiol e-pub ahead of print 25 February 2014 doi:10.1111/1462-2920.12434.
Chevin L-M, Gallet R, Gomulkiewicz R, Holt RD, Fellous S . (2013). Phenotypic plasticity in evolutionary rescue experiments. Philos Trans R Soc Lond B Biol Sci 368: 20120089.
Chevin L-M, Lande R, Mace GM . (2010). Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol 8: e1000357.
Comte J, Fauteux L, Del Giorgio PA . (2013). Links between metabolic plasticity and functional redundancy in freshwater bacterioplankton communities. Front Microbiol 4: 112.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Falgueras J, Lara A, Fernández-Pozo N, Cantón F, Pérez-Trabado G, Claros MG . (2010). SeqTrim: a high-throughput pipeline for pre-processing any type of sequence read. BMC Bioinformatics 11: 38.
Gifford SM, Sharma S, Booth M, Moran MA . (2012). Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J 7: 281–298.
Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA . (2011). Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J 5: 461–472.
Griffiths BS, Ritz K, Bardgett RD, Cook R, Christensen S, Ekelund F et al. (2000). Ecosystem response of pasture soil communities to fumigation-induced microbial diversity reductions: an examination of the biodiversity-ecosystem function relationship. Oikos 90: 279–294.
Guillard RR . (1975). Culture of phytoplankton for feeding marine invertebrates. In Smith CL, Chanley MH, (eds) Culture of Marine Invertebrate Animals. Plenum Press: New York, pp 26–60.
Jadwiszczak P . (2009). Rundom Pro 3.14. Software for classical and computer-intensive statistics http://pjadw.tripod.com.
Jansson JK, Neufeld JD, Moran MA, Gilbert JA . (2012). Omics for understanding microbial functional dynamics. Environ Microbiol 14: 1–3.
Jump AS, Penuelas J . (2005). Running to stand still: adaptation and the response of plants to rapid climate change. Ecol Lett 8: 1010–1020.
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M et al. (2007). KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484.
Landa M, Cottrell MT, Kirchman DL, Blain S, Obernosterer I . (2013a). Changes in bacterial diversity in response to dissolved organic matter supply in a continuous culture experiment. Aquat Microb Ecol 69: 157–168.
Landa M, Cottrell MT, Kirchman DL, Kaiser K, Medeiros PM, Tremblay L et al. (2013b). Phylogenetic and structural response of heterotrophic bacteria to dissolved organic matter of different chemical composition in a continuous culture study. Environ Microbiol 16: 1668–1681.
Langenheder S, Lindström ES, Tranvik LJ . (2006). Structure and function of bacterial communities emerging from different sources under identical conditions. Appl Environ Microbiol 72: 212–220.
Mantel N . (1967). Detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–230.
Moran MA, Buchan A, Gonzalez JM, Heidelberg JF, Whitman WB, Kiene RP et al. (2004). Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432: 910–913.
Mou XZ, Sun SL, Edwards RA, Hodson RE, Moran MA . (2008). Bacterial carbon processing by generalist species in the coastal ocean. Nature 451: 708–U4.
Pigliucci M . (2001) Phenotypic Plasticity Beyond Nature and Nurture. The John Hopkins University Press: Baltimore.
Polz MF, Hunt DE, Preheim SP, Weinreich DM . (2006). Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos Trans R Soc B Biol Sci 361: 2009–2021.
Rodrigue S, Materna AC, Timberlake SC, Blackburn MC, Malmstrom RR, Alm EJ et al. (2010). Unlocking short read sequencing for metagenomics. PLoS One 5: e11840.
Stewart FJ, Ottesen EA, DeLong EF . (2010). Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J 4: 896–907.
Wennersten L, Forsman A . (2012). Population-level consequences of polymorphism, plasticity and randomized phenotype switching: a review of predictions. Biol Rev Camb Philos Soc 87: 756–767.
Yachi S, Loreau M . (1999). Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci USA 96: 1463–1468.
Acknowledgements
We appreciate Shalabh Sharma, Marcelino Suzuki, Michel Krawczyk and Olivier Lagrasse for assistance with bioinformatics analyses and we appreciate Stéphane Blain for help during the experimental setup. This work was supported by grants from the Agence Nationale de la Recherche (ANR, Project BACCIO, A Biomolecular Approach to the Cycling of Carbon and Iron in the Ocean, ANR-08-BLAN-0 309) and the Gordon and Betty Moore Foundation (Grant no. 538.01).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Beier, S., Rivers, A., Moran, M. et al. Phenotypic plasticity in heterotrophic marine microbial communities in continuous cultures. ISME J 9, 1141–1151 (2015). https://doi.org/10.1038/ismej.2014.206
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2014.206
This article is cited by
-
Dynamic character displacement among a pair of bacterial phyllosphere commensals in situ
Nature Communications (2022)
-
Phenotypic plasticity of fungal traits in response to moisture and temperature
ISME Communications (2021)
-
Bivariate genome-wide association study of the growth plasticity of Staphylococcus aureus in coculture with Escherichia coli
Applied Microbiology and Biotechnology (2020)
-
Dispersal timing and drought history influence the response of bacterioplankton to drying–rewetting stress
The ISME Journal (2017)
-
Response of marine bacterioplankton pH homeostasis gene expression to elevated CO2
Nature Climate Change (2016)