Abstract
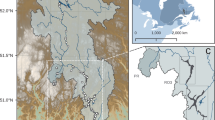
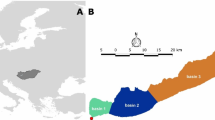
Disentangling the mechanisms shaping bacterioplankton communities across freshwater ecosystems requires considering a hydrologic dimension that can influence both dispersal and local sorting, but how the environment and hydrology interact to shape the biogeography of freshwater bacterioplankton over large spatial scales remains unexplored. Using Illumina sequencing of the 16S ribosomal RNA gene, we investigate the large-scale spatial patterns of bacterioplankton across 386 freshwater systems from seven distinct regions in boreal Québec. We show that both hydrology and local water chemistry (mostly pH) interact to shape a sequential structuring of communities from highly diverse assemblages in headwater streams toward larger rivers and lakes dominated by fewer taxa. Increases in water residence time along the hydrologic continuum were accompanied by major losses of bacterial richness and by an increased differentiation of communities driven by local conditions (pH and other related variables). This suggests that hydrology and network position modulate the relative role of environmental sorting and mass effects on community assembly by determining both the time frame for bacterial growth and the composition of the immigrant pool. The apparent low dispersal limitation (that is, the lack of influence of geographic distance on the spatial patterns observed at the taxonomic resolution used) suggests that these boreal bacterioplankton communities derive from a shared bacterial pool that enters the networks through the smallest streams, largely dominated by mass effects, and that is increasingly subjected to local sorting of species during transit along the hydrologic continuum.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Adams HE, Crump BC, Kling GW . (2014). Metacommunity dynamics of bacteria in an arctic lake: the impact of species sorting and mass effects on bacterial production and biogeography. Front Microbiol 5: 82.
Anderson MJ . (2011). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46.
Anderson MJ . (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62: 245–253.
Barberán A, Casamayor EO . (2010). Global phylogenetic community structure and β-diversity patterns in surface bacterioplankton metacommunities. Aquat Microb Ecol 59: 1–10.
Besemer K, Singer G, Quince C, Bertuzzo E, Sloan W, Battin TJ . (2013). Headwaters are critical reservoirs of microbial diversity for fluvial networks. Proc R Soc B 280: 20131760.
Borcard D, Gillet F, Legendre P . (2011) Numerical Ecology with R. Springer Science and Business Media: New York, NY, USA.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6: 1621–1624.
Cottrell MT, del Giorgio PA, Kirchman DL . (2015). Biogeography of globally distributed bacteria in temperate and boreal Québec lakes as revealed by tag pyrosequencing of 16S rRNA genes. Aquat Microb Ecol; doi: 10.3354/ame01776.
Crump BC, Adams HE, Hobbie JE, Kling GW . (2007). Biogeography of bacterioplankton in lakes and streams of an Arctic Tundra catchment. Ecology 88: 1365–1378.
Crump BC, Amaral-Zettler LA, Kling GW . (2012). Microbial diversity in arctic freshwaters is structured by inoculation of microbes from soils. ISMEJ 6: 1629–1639.
Crump BC, Hopkinson CS, Sogin ML, Hobbie JE . (2004). Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl Environ Microb 70: 1494–1505.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Fierer N, Morse JL, Berthrong ST, Bernhardt ES, Jackson RB . (2007). Environmental controls on the landscape-scale biogeography of stream bacterial communities. Ecology 88: 2162–2173.
Fierer N, Jackson RB . (2006). The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103: 626–631.
Fujii M, Kojima H, Iwata T, Urabe J, Fukui M . (2012). Dissolved organic carbon as major environmental factor affecting bacterioplankton communities in mountain lakes of Eastern Japan. Microb Ecol 63: 496–508.
Geobase . (2009). Land Cover, Circa 2000-Vector..
Gibson JJ, Prepas EE, McEachern P . (2002). Quantitative comparison of lake throughflow, residency, and catchment runoff using stable isotopes: modelling and results from a regional survey of Boreal lakes. J Hydrol 262: 128–144.
Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308.
Jones S, Cadkin T, Newton R, McMahon K . (2012). Spatial and temporal scales of aquatic bacterial beta diversity. Front Microbiol 3: 318.
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21: 494–504.
Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH . (2012). Beyond biogeographic patterns:processes shaping the microbiallandscape. Nat Rev Microbiol 10: 497–506.
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A . (2005). Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25: 1965–1978.
Heino J, Tolkkinen M, Pirttilä AM, Aisala H, Mykrä H . (2014). Microbial diversity and community– environment relationships in boreal streams. J Biogeogr 41: 2234–2244.
Langenheder S, Kely AJ . (2011). Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J 5: 1086–1094.
Lapierre JF, del Giorgio PA . (2014). Partial coupling and differential regulation of biologically and photochemically labile dissolved organic carbon across boreal aquatic networks. Biogeosciences 11: 5969–5985.
Lapierre JF, Seekell DA, del Giorgio PA . (2015). Climate and landscape influence on indicators of lake carbon cycling through spatial patterns in dissolved organic carbon. Glob Change Biol 21: 4425–4435.
Lindström ES, Bergström A-K . (2004). Influence of inlet bacteria on bacterioplankton assemblage composition in lakes of different hydraulic retention time. Limnol Oceangr 49: 125–136.
Lindström ES, Forslund M, Algesten G, Bergström A-K . (2006). External control of bacterial community structure in lakes. Limnol Oceangr 51: 339–342.
Lear G, Washington V, Neale M, Case B, Buckley H, Lewis G . (2013). The biogeography of stream bacteria. Global Ecol Biogeog 22: 544–554.
Lindström ES, Langenheder S . (2012). Local and regional factors influencing bacterial community assembly. Environ Microbiol Reports 4: 1–9.
Liu L, Yang J, Yu Z, Wilkinson D . (2015). The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J 9: 2068–2077.
Logares R, Lindström ES, Langenheder S, Logue JB, Paterson H, Laybourn-Parry J et al. (2013). Biogeography of bacterial communities exposed to progressive long-term environmental change. ISME J 7: 937–948.
Logue JB, Lindström ES . (2010). Species sorting affects bacterioplankton community composition as determined by 16 S rDNA and 16 S rRNA fingerprints. ISME J 4: 729–738.
Lozupone CA, Knight R . (2007). Global patterns in bacterial diversity. Proc Natl Acad Sci USA 104: 11436–11440.
Magoč T, Salzberg SL . (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963.
Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4: 102–112.
Nelson CE, Sadro S, Melack JM . (2009). Contrasting the influences of stream inputs and landscape position on bacterioplankton community structure and dissolved organic matter composition in high‐elevation lake chains. Limnol Oceangr 54: 1292–1305.
Newton RJ, Jones SE, Eiler A, McMahon KA, Bertilsson S . (2011). A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol R 75: 14–49.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MH, Wagner H . (2015), vegan: Community Ecology Package. R package version 2.3-0 http://CRAN.R-project.org/package=vegan.
Pommier T, Canbäck B, Riemann L, Boström KH, Simu K, Lundberg P et al. (2006). Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol 16: 867–880.
R Core Team . (2013). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. Available from http://www.R-project.org/.
Read D, Gweon H, Bowes M, Newbold L, Field D, Bailey M, Griffiths R . (2015). Catchment-scale biogeography of riverine bacterioplankton. ISME J 9: 516–526.
Ren L, Jeppesen E, He D, Wang J, Liboriussen L, Xing P et al. (2015). pH influences the importance of niche-related and neutral processes in lacustrine bacterioplankton assembly. Wommack, KE (ed). Appl Environ Microb 81: 3104–3114.
Rasilo T, Prairie YT, del Giorgio PA . (2015). Large-scale patterns in summer diffusive CH4 fluxes across boreal lakes, and contribution to diffusive C emissions. Glob Change Biol 21: 1124–1139.
Ruiz-González C, Niño-García JP, Lapierre JF, del Giorgio PA . (2015a). The quality of organic matter shapes the functional biogeography of bacterioplankton across boreal freshwater ecosystems. Global Ecol Biogeogr 24: 1487–1498.
Ruiz-González C, Niño-García JP, del Giorgio PA . (2015b). Terrestrial origin bacterial communities in complex boreal freshwater networks. Ecol Lett 18: 1198–1206.
Savio D, Sinclair L, Ijaz UZ, Blaschke AP, Reischer GH, Bloeschl G et al. (2015). Bacterial diversity along a 2 600 km river continuum. Environ Microbiol; doi: 10.1111/1462-2920.12886.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb 75: 7537–7541.
Shade A, Caporaso JG, Handelsman J, Knight R, Fierer N . (2013). A meta-analysis of changes in bacterial and archaeal communities with time. ISME J 7: 1493–1506.
Souffreau C, Van der Gucht K, Van Gremberghe I, Kosten S, Lacerot G, Meirelles Lobão L et al. (2015). Environmental rather than spatial factors structure bacterioplankton communities in shallow lakes along a > 6000 km latitudinal gradient in South America. Environ Microbiol 17: 2336–2351.
Székely AJ, Berga ME, Langenheder S . (2012). Mechanisms determining the fate of dispersed bacterial communities in new environments. ISME J 7: 61–71.
Turner KW, Edwards TWD, Wolfe BB . (2014). Characterising runoff generation processes in a Lake-Rich Thermokarst Landscape (Old Crow Flats, Yukon, Canada) using δ 18O, δ 2H and d-excess measurements. Permafrost Periglac 25: 53–59.
Van der Gucht K, Cottenie K, Muylaert K, Vloemans N, Cousin S, Declerck S et al. (2007). The power of species sorting: local factors drive bacterial community composition over a wide range of spatial scales. Proc Natl Acad Sci USA 104: 20404–20409.
Wang Q, Garrity GM, Tiedje JM, Cole JR . (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb 73: 5261–5267.
Wilhelm L, Besemer K, Fragner L, Peter H, Weckwerth W, Battin T . (2015). Altitudinal patterns of diversity and functional traits of metabolically active microorganisms in stream biofilms. ISME J 9: 2454–2464.
Yannarell AC, Triplett EW . (2005). Geographic and environmental sources of variation in lake bacterial community composition. Appl Environ Microb 71: 227–239.
Acknowledgements
We thank Ryan Hutchins for help with GIS analyses, Annick St-Pierre, Alice Parkes and the whole CarBBAS team for their contribution to the field and laboratory components of this research, and Nicolas Fortin St-Gelais for critical comments and discussion. This study is part of the program of the Carbon Biogeochemistry in Boreal Aquatic Systems (CarBBAS) Industrial Research Chair, co-funded by the Natural Science and Engineering Research Council of Canada (NSERC) and Hydro-Québec.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Niño-García, J., Ruiz-González, C. & del Giorgio, P. Interactions between hydrology and water chemistry shape bacterioplankton biogeography across boreal freshwater networks. ISME J 10, 1755–1766 (2016). https://doi.org/10.1038/ismej.2015.226
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2015.226
This article is cited by
-
High-resolution freshwater dissolved calcium and pH data layers for Canada and the United States
Scientific Data (2024)
-
Effects of Cascade Reservoirs on Spatiotemporal Dynamics of the Sedimentary Bacterial Community: Co-occurrence Patterns, Assembly Mechanisms, and Potential Functions
Microbial Ecology (2024)
-
The different responses of planktonic bacteria and archaea to water temperature maintain the stability of their community diversity in dammed rivers
Ecological Processes (2023)
-
Glacial recession in Andean North-Patagonia (Argentina): microbial communities in benthic biofilms of glacier-fed streams
Hydrobiologia (2023)
-
Spatial and Seasonal Patterns of Sediment Bacterial Communities in Large River Cascade Reservoirs: Drivers, Assembly Processes, and Co-occurrence Relationship
Microbial Ecology (2023)