Abstract
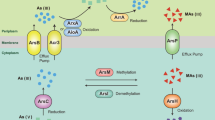
The globally significant picocyanobacterium Prochlorococcus is the main primary producer in oligotrophic subtropical gyres. When phosphate concentrations are very low in the marine environment, the mol:mol availability of phosphate relative to the chemically similar arsenate molecule is reduced, potentially resulting in increased cellular arsenic exposure. To mediate accidental arsenate uptake, some Prochlorococcus isolates contain genes encoding a full or partial efflux detoxification pathway, consisting of an arsenate reductase (arsC), an arsenite-specific efflux pump (acr3) and an arsenic-related repressive regulator (arsR). This efflux pathway was the only previously known arsenic detox pathway in Prochlorococcus. We have identified an additional putative arsenic mediation strategy in Prochlorococcus driven by the enzyme arsenite S-adenosylmethionine methyltransferase (ArsM) which can convert inorganic arsenic into more innocuous organic forms and appears to be a more widespread mode of detoxification. We used a phylogenetically informed approach to identify Prochlorococcus linked arsenic genes from both pathways in the Global Ocean Sampling survey. The putative arsenic methylation pathway is nearly ubiquitously present in global Prochlorococcus populations. In contrast, the complete efflux pathway is only maintained in populations which experience extremely low PO4:AsO4, such as regions in the tropical and subtropical Atlantic. Thus, environmental exposure to arsenic appears to select for maintenance of the efflux detoxification pathway in Prochlorococcus. The differential distribution of these two pathways has implications for global arsenic cycling, as their associated end products, arsenite or organoarsenicals, have differing biochemical activities and residence times.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Abascal F, Zardoya R, Telford MJ . (2010). TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38: W7–W13.
Ajees AA, Marapakala K, Packianathan C, Sankaran B, Rosen BP . (2012). Structure of an As(III) S-adenosylmethionine methyltransferase: insights into the mechanism of arsenic biotransformation. Biochemistry 51: 5476–5485.
Akter KF, Owens G, Davey DE, Naidu R . (2005). Arsenic speciation and toxicity in biological systems. Rev Environ Contam Toxicol 184: 97–149.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Andreae MO . (1979). Arsenic speciation in seawater and interstitial waters: the influence of biological-chemical interactions on the chemistry of a trace element. Limnol Oceanogr 24: 440–452.
Bennett MS, Guan Z, Laurberg M, Su XD . (2001). Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases. Proc Natl Acad Sci USA 98: 13577–13582.
Berger SA, Krompass D, Stamatakis A . (2011). Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood. Syst Biol 60: 291–302.
Berger SA, Stamatakis A . (2011). Aligning short reads to reference alignments and trees. Bioinformatics 27: 2068–2075.
Bertilsson S, Berglund O, Karl DM, Chisholm SW . (2003). Elemental composition of marine Prochlorococcus and Synechococcus: Implications for the ecological stoichiometry of the sea. Limnol Oceanogr 48: 1721–1731.
Campbell L, Liu H, Nolla HA, Vaulot D . (1997). Annual variability of phytoplankton and bacteria in the subtropical North Pacific Ocean at Station ALOHA during the 1991–1994 ENSO event. Deep Sea Research Part I: Oceanographic Research Papers 44: 167–192.
Campbell L, Landry MR, Constantinou J, Nolla HA, Brown SL, Liu H et al. (1998). Response of microbial community structure to environmental forcing in the Arabian Sea. Deep Sea Research Part II: Topical Studies in Oceanography 45: 2301–2325.
Carlin A, Shi W, Dey S, Rosen BP . (1995). The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol 177: 981–986.
Chen F, Mackey AJ, Stoeckert CJ, Roos DS . (2006). OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res 34: D363–D368.
Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, DeLong EF et al. (2006). Genomic islands and the ecology and evolution of Prochlorococcus. Science 311: 1768–1770.
Coleman ML, Chisholm SW . (2010). Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA 107: 18634–18639.
Cutter GA . (1992). Kinetic controls on metalloid speciation in seawater. Mar Chem 40: 65–80.
Cutter GA, Cutter LS . (1998). Metalloids in the high latitude North Atlantic Ocean: Sources and internal cycling. Mar Chem 61: 25–36.
Cutter GA, Cutter LS, Featherstone AM, Lohrenz SE . (2001). Antimony and arsenic biogeochemistry in the western Atlantic Ocean. Deep Sea Research Part II: Topical Studies in Oceanography 48: 2895–2915.
Cutter GA, Cutter LS . (2006). Biogeochemistry of arsenic and antimony in the North Pacific Ocean. Geochemistry Geophysics Geosystems 7: 1–12.
Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D et al. (2009). MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res 38: D396–D400.
DeLong EF . (2005). Microbial community genomics in the ocean. Nat Rev Microbiol 3: 459–469.
Dembitsky VM, Levitsky DO . (2004). Arsenolipids. Prog Lipid Res 43: 403–448.
DuRand MD, Olson RJ, Chisholm SW . (2001). Phytoplankton population dynamics at the Bermuda Atlantic Time-series station in the Sargasso Sea. Deep Sea Research Part II: Topical Studies in Oceanography 48: 1983–2003.
Dyhrman ST, Haley ST . (2011). Arsenate resistance in the unicellular marine diazotroph Crocosphaera watsonii. Front Microbiol 2: 214.
Elias M, Wellner A, Goldin-Azulay K, Chabriere E, Vorholt JA, Erb TJ et al. (2012). The molecular basis of phosphate discrimination in arsenate-rich environments. Nature 491: 134–137.
Ellwood MJ, Maher WA . (2002). An automated hydride generation-cryogenic trapping-ICP-MS system for measuring inorganic and methylated Ge, Sb and As species in marine and fresh waters. J Anal At Spectrom 17: 197–203.
Feldmann J, Krupp EM . (2011). Critical review or scientific opinion paper: arsenosugars—a class of benign arsenic species or justification for developing partly speciated arsenic fractionation in foodstuffs? Anal Bioanal Chem 399: 1735–1741.
Garcia HE, Locarnini RA, Boyer TP, Antonov JI, Zweng MM, Baranova OK et al. (2010) World Ocean Atlas 2009, Volume 4: Nutrients (phosphate, nitrate, silicate). NOAA Atlas NESDIS 71. U.S. Government Printing Office: Washington, D.C, p 398.
Ghosh M, Shen J, Rosen BP . (1999). Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 96: 5001–5006.
Harrington CF, Brima EI, Jenkins RO . (2008). Biotransformation of arsenobetaine by microorganisms from the human gastrointestinal tract. Chemical Speciation and Bioavailability 20: 173–180.
Heldal M, Scanlan DJ, Norland S, Thingstad F, Mann NH . (2003). Elemental composition of single cells of various strains of marine Prochlorococcus and Synechococcus using X-ray microanalysis. Limnol Oceanogr 48: 1732–1743.
Hewson I, Poretsky RS, Dyhrman ST, Zielinski B, White AE, Tripp HJ et al. (2009). Microbial community gene expression within colonies of the diazotroph, Trichodesmium, from the Southwest Pacific Ocean. ISME J 3: 1286–1300.
Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EM, Chisholm SW . (2006). Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311: 1737–1740.
Kaneko T, Nakamura Y, Sasamoto S, Watanabe A, Kohara M, Matsumoto M et al. (2003). Structural analysis of four large plasmids harboring in a unicellular cyanobacterium, Synechocystis sp. PCC 6803. DNA Res 10: 221–228.
Karl DM, Tien G . (1997). Temporal variability in dissolved phosphorus concentrations in the subtropical North Pacific Ocean. Mar Chem 56: 77–96.
Karl DM . (2007) The Marine Phosphorus Cycle. ASM Press: Washington, pp 523–539.
Kelley LA, Sternberg MJE . (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363–371.
Kelly L, Ding HM, Huang KH, Osburne MS, Chisholm SW . (2013). Genetic diversity in cultured and wild marine cyanomyoviruses reveals phosphorus stress as a strong selective agent. ISME J 7: 1827–1841.
Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S et al. (2007). Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet 3: e231.
Krumhardt KM, Callnan K, Roache-Johnson K, Swett T, Robinson D, Reistetter EN et al. (2013). Effects of phosphorus starvation versus limitation on the marine cyanobacterium Prochlorococcus MED4 I: uptake physiology. Environ Microbiol 15: 2114–2128.
Langmead B, Salzberg SL . (2012). Fast gapped-read alignment with Bowtie 2. Nat Meth 9: 357–359.
Leffers L, Ebert F, Taleshi MS, Francesconi KA, Schwerdtle T . (2013a). In vitro toxicological characterization of two arsenosugars and their metabolites. Mol Nutr Food Res 57: 1270–1282.
Leffers L, Wehe CA, Huwel S, Bartel M, Ebert F, Taleshi MS et al. (2013b). In vitro intestinal bioavailability of arsenosugar metabolites and presystemic metabolism of thio-dimethylarsinic acid in Caco-2 cells. Metallomics 5: 1031–1042.
Lunde G . (1973). The synthesis of fat and water soluble arseno organic compounds in marine and limnetic algae. Acta Chem Scand 27: 1586–1594.
Mandal BK, Suzuki KT . (2002). Arsenic round the world: a review. Talanta 58: 201–235.
Martiny AC, Coleman ML, Chisholm SW . (2006). Phosphate acquisition genes in Prochlorococcus ecotypes: Evidence for genome-wide adaptation. Proc Natl Acad Sci USA 103: 12552–12557.
Martiny AC, Huang Y, Li W . (2009). Occurrence of phosphate acquisition genes inProchlorococcuscells from different ocean regions. Environ Microbiol 11: 1340–1347.
Meyer S, Matissek M, Muller SM, Taleshi MS, Ebert F, Francesconi KA et al. (2014). In vitro toxicological characterisation of three arsenic-containing hydrocarbons. Metallomics 6: 1023–1033.
Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L et al. (2013). Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14: 193–202.
Miyashita S-I, Fujiwara S, Tsuzuki M, Kaise T . (2012). Cyanobacteria produce arsenosugars. Environmental Chemistry 9: 474–484.
Moore LR, Rocap G, Chisholm SW . (1998). Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393: 464–467.
Moore LR, Chisholm SW . (1999). Photophysiology of the Marine Cyanobacterium Prochlorococcus: Ecotypic Differences among Cultured Isolates. Limnol Oceanogr 44: 628–638.
Moore LR, Anton FP, Rocap G, Chisholm SW . (2002). Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol Oceanogr 47: 989–996.
Mukhopadhyay R, Rosen BP, Phung LT, Silver S . (2002). Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol Rev 26: 311–325.
Newcombe C, Raab A, Williams PN, Deacon C, Haris PI, Meharg AA et al. (2010). Accumulation or production of arsenobetaine in humans? J Environ Monit 12: 832–837.
Oremland RS, Stolz JF . (2003). The ecology of arsenic. Science 300: 939–944.
Páez-Espino D, Tamames J, Lorenzo V, Cánovas D . (2009). Microbial responses to environmental arsenic. BioMetals 22: 117–130.
Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C . (2006). Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci USA 103: 2075–2080.
Raml R, Raber G, Rumpler A, Bauernhofer T, Goessler W, Francesconi KA . (2009). Individual variability in the human metabolism of an arsenic-containing carbohydrate, 2',3'-dihydroxypropyl 5-deoxy-5-dimethylarsinoyl-beta-D-riboside, a naturally occurring arsenical in seafood. Chem Res Toxicol 22: 1534–1540.
Reistetter EN, Krumhardt K, Callnan K, Roache-Johnson K, Saunders JK, Moore LR et al. (2013). Effects of phosphorus starvation versus limitation on the marine cyanobacterium Prochlorococcus MED4 II: gene expression. Environ Microbiol 15: 2129–2143.
Rocap G, Distel DL, Waterbury JB, Chisholm SW . (2002). Resolution of Prochlorococcus and Synechococcus ecotypes by using 16 S-23 S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol 68: 1180–1191.
Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA et al. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424: 1042–1047.
Rosen BP . (2002). Biochemistry of arsenic detoxification. FEBS Lett 529: 86–92.
Rosenberg H, Gerdes RG, Chegwidden K . (1977). Two systems for the uptake of phosphate in Escherichia coli. J Bacteriol 131: 505–511.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling Expedition: northwest Atlantic through Eastern tropical Pacific. PLoS Biol 5: e77.
Scanlan D . (2012) Marine Picocyanobacteria. In: Whitton BA (ed). Ecology of Cyanobacteria II. Springer: Netherlands, pp 503–533.
Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR et al. (2009). Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73: 249–299.
Stamatakis A . (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690.
Stolz JF, Basu P, Santini JM, Oremland RS . (2006). Arsenic and selenium in microbial metabolism. Annu Rev Microbiol 60: 107–130.
Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW . (2005). Three Prochlorococcus cyanophage genomes: Signature features and ecological interpretations. PLoS Biol 3: 790–806.
Sun S, Chen J, Li W, Altintas I, Lin A, Peltier S et al. (2011). Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: the CAMERA resource. Nucleic Acids Res 39: D546–D551.
Tatusov RL, Galperin MY, Natale DA, Koonin EV . (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28: 33–36.
Tawfik DS, Viola RE . (2011). Arsenate replacing phosphate: alternative life chemistries and ion promiscuity. Biochemistry 50: 1128–1134.
Thomas DJ, Nava GM, Cai SY, Boyer JL, Hernandez-Zavala A, Gaskins HR . (2010). Arsenic (+3 oxidation state) methyltransferase and the methylation of arsenicals in the invertebrate chordate Ciona intestinalis. Toxicol Sci 113: 70–76.
Van Mooy BAS, Rocap G, Fredricks HF, Evans CT, Devol AH . (2006). Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marine environments. Proc Natl Acad Sciences USA 103: 8607–8612.
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74.
Ventura-Lima J, Bogo MR, Monserrat JM . (2011). Arsenic toxicity in mammals and aquatic animals: A comparative biochemical approach. Ecotoxicol Environ Saf 74: 211–218.
Wass MN, Kelley LA, Sternberg MJ . (2010). 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res 38: W469–W473.
Wu J, Sunda W, Boyle EA, Karl DM . (2000). Phosphate depletion in the western North Atlantic Ocean. Science 289: 759–762.
Wurl O, Zimmer L, Cutter GA . (2013). Arsenic and phosphorus biogeochemistry in the ocean: Arsenic species as proxies for P-limitation. Limnol Oceanogr 58: 729–740.
Xu C, Shi W, Rosen BP . (1996). The chromosomal arsR gene of Escherichia coli encodes a trans-acting metalloregulatory protein. J Biol Chem 271: 2427–2432.
Yin XX, Chen J, Qin J, Sun GX, Rosen BP, Zhu YG . (2011). Biotransformation and volatilization of arsenic by three photosynthetic cyanobacteria. Plant Physiol 156: 1631–1638.
Yuan Y, Barry CE 3rd . (1996). A common mechanism for the biosynthesis of methoxy and cyclopropyl mycolic acids in Mycobacterium tuberculosis. Proc Natl Acad Sci USA 93: 12828–12833.
Acknowledgements
We thank Cedar McKay for help setting up local computational resources. We would also like to thank Simon Berger for providing amino acid support to the PaPaRa alignment program. This work was made possible through support from a National Science Foundation Graduate Research Fellowship to JKS and NSF OCE-1138368 to GR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Saunders, J., Rocap, G. Genomic potential for arsenic efflux and methylation varies among global Prochlorococcus populations. ISME J 10, 197–209 (2016). https://doi.org/10.1038/ismej.2015.85
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2015.85
This article is cited by
-
Acclimation and adaptation to elevated pCO2 increase arsenic resilience in marine diatoms
The ISME Journal (2021)
-
Biological characterization of Bacillus flexus strain SSAI1 transforming highly toxic arsenite to less toxic arsenate mediated by periplasmic arsenite oxidase enzyme encoded by aioAB genes
BioMetals (2021)
-
Unlocking the Genomic Taxonomy of the Prochlorococcus Collective
Microbial Ecology (2020)
-
Cyanobacteria and cyanophage contributions to carbon and nitrogen cycling in an oligotrophic oxygen-deficient zone
The ISME Journal (2019)
-
Two facets of world arsenic problem solution: crop poisoning restriction and enforcement of phytoremediation
Planta (2018)