Abstract
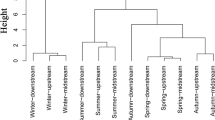
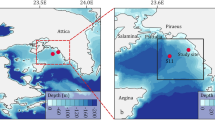
Mycoplankton have so far been a neglected component of pelagic marine ecosystems, having been poorly studied relative to other plankton groups. Currently, there is a lack of understanding of how mycoplankton diversity changes through time, and the identity of controlling environmental drivers. Using Fungi-specific high-throughput sequencing and quantitative PCR analysis of plankton DNA samples collected over 6 years from the coastal biodiversity time series site Station L4 situated off Plymouth (UK), we have assessed changes in the temporal variability of mycoplankton diversity and abundance in relation to co-occurring environmental variables. Mycoplankton diversity at Station L4 was dominated by Ascomycota, Basidiomycota and Chytridiomycota, with several orders within these phyla frequently abundant and dominant in multiple years. Repeating interannual mycoplankton blooms were linked to potential controlling environmental drivers, including nitrogen availability and temperature. Specific relationships between mycoplankton and other plankton groups were also identified, with seasonal chytrid blooms matching diatom blooms in consecutive years. Mycoplankton α-diversity was greatest during periods of reduced salinity at Station L4, indicative of riverine input to the ecosystem. Mycoplankton abundance also increased during periods of reduced salinity, and when potential substrate availability was increased, including particulate organic matter. This study has identified possible controlling environmental drivers of mycoplankton diversity and abundance in a coastal sea ecosystem, and therefore sheds new light on the biology and ecology of an enigmatic marine plankton group. Mycoplankton have several potential functional roles, including saprotrophs and parasites, that should now be considered within the consensus view of pelagic ecosystem functioning and services.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Abarenkov K, Henrik Nilsson R, Larsson K, Alexander I, Eberhardt U, Erland S et al. (2010). The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol 189: 281–285.
Bass D, Howe A, Brown N, Barton H, Demidova M, Michelle H et al. (2007). Yeast forms dominate fungal diversity in the deep oceans. Proc R Soc B 274: 3069–3077.
Brewer PG, Riley JP . (1965). The automatic determination of nitrate in sea water. Deep-Sea Res 12: 765–772.
Burgaud G, Woehlke S, Rédou V, Orsi W, Beaudoin D, Barbier G et al. (2013). Deciphering the presence and activity of fungal communities in marine sediments using a model estuarine system. Aquat Microb Ecol 70: 45–62.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA . (2012). The Western English Channel contains a persistent microbial seed bank. ISME J 6: 1089–1093.
de Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 348: 1261605.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461.
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R . (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200.
Edgar RC . (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998.
Eloire D, Somerfield PJ, Conway DVP, Halsband-Lenk C, Harris R, Bonnet D . (2010). Temporal variability and community composition of zooplankton at station L4 in the Western Channel: 20 years of sampling. J Plankton Res 32: 657–679.
Fuhrman JA, Cram JA, Needham DM . (2015). Marine microbial community dynamics and their ecological interpretation. Nat Rev Microbiol 13: 133–146.
Gao Z, Johnson ZI, Wang G . (2010). Molecular characterization of the spatial diversity and novel lineages of mycoplankton in Hawaiian coastal waters. ISME J 4: 111–120.
Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T et al. (2009). The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol 11: 3132–3139.
Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308.
Gleason FH, Küpper FC, Amon JP, Picard K, Gachon CMM, Marano AV et al. (2011). Zoosporic true fungi in marine ecosystems: a review. Mar Freshw Res 62: 383–393.
Grasshoff K, Kremling K, Ehrhardt M, Anderson LG . (1999) Methods of Seawater Analysis. Wiley-VCH: Weinheim.
Gsell AS, de Senerpont Domis LN, Verhoeven KJ, van Donk E, Ibelings BW . (2013). Chytrid epidemics may increase genetic diversity of a diatom spring-bloom. ISME J 7: 2057–2059.
Gutiérrez MH, Pantoja S, Tejos E, Quiñones RA . (2011). The role of fungi in processing marine organic matter in the upwelling ecosystem off Chile. Mar Biol 158: 205–219.
Kagami M, Bruin Ad, Ibelings BW, Donk EV . (2007). Parasitic chytrids: their effects on phytoplankton communities and food-web dynamics. Hydrobiologia 578: 113–129.
Karl DM, Church MJ . (2014). Microbial oceanography and the Hawaii Ocean Time-series programme. Nat Rev Microbiol 12: 699–713.
Kirkwood DS . (1989). Simultaneous determination of selected nutrients in seawater. ICES CM C:29, 12.
Lepelletier F, Karpov SA, Alacid E, Le Panse S, Bigeard E, Garcés E et al. (2014). Dinomyces arenysensis gen. et sp. nov. (Rhizophydiales, Dinomycetaceae fam. nov.), a chytrid infecting marine dinoflagellates. Protist 165: 230–244.
Logares R, Audic S, Bass D, Bittner L, Boutte C, Christen R et al. (2014). Patterns of rare and abundant marine microbial eukaryotes. Curr Biol 24: 813–821.
Manohar CS, Raghukumar C . (2013). Fungal diversity from various marine habitats deduced through culture-independent studies. FEMS Microbiol Lett 341: 69–78.
Mantoura RFC, Woodward EMS . (1983). Optimization of the indophenol blue method for the automated determination of ammonia in estuarine waters. Estuar Coast Sheld Sea Sci 17: 219–224.
Mary I, Cummings DG, Biegala IC, Burkill PH, Archer SD, Zubkov MV . (2006). Seasonal dynamics of bacterioplankton community structure at a coastal station in the western English Channel. Aquat Microb Ecol 42: 119–126.
Mohamed DJ, Martiny JB . (2011). Patterns of fungal diversity and composition along a salinity gradient. ISME J 5: 379–388.
Orsi W, Biddle JF, Edgcomb V . (2013a). Deep sequencing of subseafloor eukaryotic rRNA reveals active Fungi across marine subsurface provinces. PLoS One 8: e56335.
Orsi WD, Edgcomb VP, Christman GD, Biddle JF . (2013b). Gene expression in the deep biosphere. Nature 499: 205–210.
Peacock EE, Olson RJ, Sosik HM . (2014). Parasitic infection of the diatom Guinardia delicatula, a recurrent and ecologically important phenomenon on the New England Shelf. Mar Ecol Prog Ser 503: 1–10.
Pohnert G, Steinke M, Tollrian R . (2007). Chemical cues, defence metabolites and the shaping of pelagic interspecific interactions. Trends Ecol Evol 22: 198–204.
Prevost-Boure NC, Christen R, Dequiedt S, Mougel C, Lelievre M, Jolivet C et al. (2011). Validation and application of a PCR primer set to quantify fungal communities in the soil environment by real-time quantitative PCR. PLoS One 6: e24166.
Richards TA, Jones MDM, Leonard G, Bass D . (2012). Marine fungi: their ecology and molecular diversity. Annu Rev Marine Sci 4: 495–522.
Russell FS, Southward AJ, Boalch GT, Butler EI . (1971). Changes in biological conditions in the English Channel off Plymouth during the last half century. Nature 234: 468–470.
Santamaria M, Fosso B, Consiglio A, De Caro G, Grillo G, Licciulli F et al. (2012). Reference databases for taxonomic assignment in metagenomics. Brief Bioinform 13: 682–695.
Scholz B, Guillou L, Marano AV, Neuhauser S, Sullivan BK, Karsten U et al. (2016). Zoosporic parasites infecting marine diatoms – a black box that needs to be opened. Fungal Ecol 19: 59–76.
Smyth TJ, Fishwick JR, AL-Moosawi L, Cummings DG, Harris C, Kitidis V et al. (2010). A broad spatio-temporal view of the Western English Channel observatory. J Plankt Res 32: 585–601.
Southward AJ . (1974). Changes in the plankton community of the Western English Channel. Nature 249: 180–181.
Spatafora JW, Volkmann-Kohlmeyer B, Kohlmeyer J . (1998). Independent terrestrial origins of the Halosphaeriales (marine Ascomycota). Am J Bot 85: 1569–1580.
Suetrong S, Schoch CL, Spatafora JW, Kohlmeyer J, Volkmann-Kohlmeyer B, Sakayaroj J et al. (2009). Molecular systematics of the marine Dothideomycetes. Stud Mycol 64: 155–173.
Taylor DL, Houston S . (2011). A bioinformatics pipeline for sequence-based analyses of fungal biodiversity. In: Xu J-R, Bluhm BH (eds), Fungal Genomics. Springer: Berlin, Germany, pp 141–155.
Taylor JD, Cottingham SD, Billinge J, Cunliffe M . (2014). Seasonal microbial community dynamics correlate with phytoplankton-derived polysaccharides in surface coastal waters. ISME J 8: 245–248.
Taylor JD, Cunliffe M . (2014). High-throughput sequencing reveals neustonic and planktonic microbial eukaryote diversity in coastal waters. J Phycol 50: 960–965.
Taylor JD, Cunliffe M . (2015). Polychaete burrows harbour distinct microbial communities in oil-contaminated coastal sediments. Environ Microbiol Rep 7: 606–613.
Tillmann U, Hesse K-J, Tillmann A . (1999). Large-scale parasitic infection of diatoms in the Northfrisian Wadden Sea. J Sea Res 42: 251–261.
Vishniac HS . (1956). On the ecology of the lower marine fungi. Biol Bull 111: 410–414.
Wang G, Wang X, Liu X, Li Q . (2012) Diversity and biogeochemical function of planktonic fungi in the ocean. In: Raghukumar C (ed), Biology of Marine Fungi. Spinger: Berlin, Germany, pp 71–88.
Wang X, Singh P, Gao Z, Zhang X, Johnson ZI, Wang G . (2014). Distribution and diversity of planktonic fungi in the West Pacific Warm Pool. PLOS One 9: e101523.
Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F . (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol 9: 2707–2719.
Welschmeyer NA . (1994). Fluorometric analysis of chlorophyll a in the presence of chlorophyll b and pheopigments. Limnol Oceanogr 39: 1985–1992.
White TJ, Bruns T, Lee S, Taylor J . (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds), PCR Protocols: A Guide to Methods and Applications. Academic Press: San Diego, USA.
Widdicombe CE, Eloire D, Harbour D, Harris RP, Somerfield PJ . (2010). Long-term phytoplankton community dynamics in the Western English Channel. J Plankt Res 32: 643–655.
Worden AZ, Follows MJ, Giovannoni SJ, Wilken S, Zimmerman AE, Keeling PJ . (2015). Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347: 1257594.
Wurzbacher CM, Bärlocher F, Grossart HP . (2010). Fungi in lake ecosystems. Aquat Microb Ecol 59: 125–149.
Xu W, Pang KL, Luo ZH . (2014). High fungal diversity and abundance recovered in the deep-sea sediments of the Pacific Ocean. Microb Ecol 68: 688–698.
Zhang JZ, Chi J . (2002). Automated analysis of nanomolar concentrations of phosphate in natural waters with liquid waveguides. Environ Sci Technol 36: 1048–1053.
Acknowledgements
This study was funded by a Marine Biological Association (MBA) Research Fellowship awarded to MC. We thank Declan Schroeder and Cecilia Balestreri (MBA) for kindly providing the plankton genomic DNA samples used in this study from the EU FP7-funded project Marine Microbial Biodiversity, Bioinformatics, Biotechnology (MicroB3). We are indebted to the crews of RV Plymouth Quest and RV Sepia for continued sampling at Station L4. Supporting environmental metadata were provided by the Western Channel Observatory that is funded as part of the UK Natural Environment Research Council’s National Capability. Special thanks to Claire Widdicombe from the Plymouth Marine Laboratory for providing the diatom abundance and diversity data. We also thank three anonymous reviewers for helping to improve on a previous version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Taylor, J., Cunliffe, M. Multi-year assessment of coastal planktonic fungi reveals environmental drivers of diversity and abundance. ISME J 10, 2118–2128 (2016). https://doi.org/10.1038/ismej.2016.24
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2016.24
This article is cited by
-
Fungal parasitism on diatoms alters formation and bio–physical properties of sinking aggregates
Communications Biology (2023)
-
Impact of environmental factors on diversity of fungi in sediments from the Shenzhen River Estuary
Archives of Microbiology (2023)
-
Abundant and Rare Taxa of Planktonic Fungal Community Exhibit Distinct Assembly Patterns Along Coastal Eutrophication Gradient
Microbial Ecology (2023)
-
Intercomparison of Two Fluorescent Dyes to Visualize Parasitic Fungi (Chytridiomycota) on Phytoplankton
Microbial Ecology (2023)
-
Global contribution of pelagic fungi to protein degradation in the ocean
Microbiome (2022)