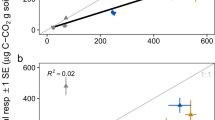
Abstract
Microorganisms perform most decomposition on Earth, mediating carbon (C) loss from ecosystems, and thereby influencing climate. Yet, how variation in the identity and composition of microbial communities influences ecosystem C balance is far from clear. Using quantitative stable isotope probing of DNA, we show how individual bacterial taxa influence soil C cycling following the addition of labile C (glucose). Specifically, we show that increased decomposition of soil C in response to added glucose (positive priming) occurs as a phylogenetically diverse group of taxa, accounting for a large proportion of the bacterial community, shift toward additional soil C use for growth. Our findings suggest that many microbial taxa exhibit C use plasticity, as most taxa altered their use of glucose and soil organic matter depending upon environmental conditions. In contrast, bacteria that exhibit other responses to glucose (reduced growth or reliance on glucose for additional growth) clustered strongly by phylogeny. These results suggest that positive priming is likely the prototypical response of bacteria to sustained labile C addition, consistent with the widespread occurrence of the positive priming effect in nature.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Ainsworth EA, Long SP . (2005). What have we learned from 15 years of free‐air CO2 enrichment (FACE)? A meta‐analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2 . New Phytol 165: 351–372.
Blagodatskaya E, Kuzyakov Y . (2008). Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: critical review. Biol Fert Soils 45: 115–131.
Blagodatskaya E, Kuzyakov Y . (2013). Active microorganisms in soil: critical review of estimation criteria and approaches. Soil Biol Biochem 67: 192–211.
Blagodatskaya EV, Blagodatsky SA, Anderson TH, Kuzyakov Y . (2007). Priming effects in chernozem induced by glucose and N in relation to microbial growth strategies. Appl Soil Ecol 37: 95–105.
Blankinship JC, Brown JR, Dijkstra P, Allwright MC, Hungate BA . (2010). Response of terrestrial CH4 uptake to interactive changes in precipitation and temperature along a climatic gradient. Ecosystems 13: 1157–1170.
Buchkowski RW, Schmitz OJ, Bradford MA . (2015). Microbial stoichiometry overrides biomass as a regulator of soil carbon and nitrogen cycling. Ecology 96: 1139–1149.
Cadotte MW, Cavender-Bares J, Tilman D, Oakley TH . (2009). Using phylogenetic, functional and trait diversity to understand patterns of plant community productivity. PLoS One 4: e5695.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high throughput community sequencing data. Nat Methods 7: 335–336.
Chen R, Senbayram M, Blagodatsky S, Myachina O, Dittert K, Lin X et al. (2014). Soil C and N availability determine the priming effect: microbial N mining and stoichiometric decomposition theories. Glob Change Biol 20: 2356–2367.
Cleveland CC, Nemergut DR, Schmidt SK, Townsend AR . (2007). Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition. Biogeochemistry 82: 229–240.
DeAngelis KM, Brodie EL, DeSantis TZ, Anderson GL, Lindow SE, Firestone MK . (2009). Selective progressive response of soil microbial community to wild oat roots. ISME J 3: 168–178.
Dijkstra P, Ishizu A, Doucett R, Hart SC, Schwartz E, Menyailo OV et al. (2006). 13C and 15N natural abundance of the soil microbial biomass. Soil Biol Biochem 38: 3257–3266.
Evans SE, Wallenstein MD . (2014). Climate change alters ecological strategies of soil bacteria. Ecol Lett 17: 155–164.
Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM et al. (2014). An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2: 6.
Fierer N, Bradford MA, Jackson RB . (2007). Toward an ecological classification of soil bacteria. Ecology 88: 1354–1364.
Fontaine S, Barot S . (2005). Size and functional diversity of microbe populations control plant persistence and long‐term soil carbon accumulation. Ecol Lett 8: 1075–1087.
Fontaine S, Mariotti A, Abbadie L . (2003). The priming effect of organic matter: a question of microbial competition? Soil Biol Biochem 35: 837–843.
Germida JJ, Siciliano SD, Seib AM . (1998). Phenotypic plasticity of Pseudomonas aureofaciens (lacZY) introduced into and recovered from field and laboratory microcosm soils. FEMS Microbiol Ecol 27: 133–139.
Goldfarb KC, Karoz U, Hanson CA, Santee CA, Bradford MA, Treseder KK et al. (2011). Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front Microbiol 2: 94.
Haei M, Rousk J, Ilstedt U, Öquist M, Bååth E, Laudon H . (2011). Effects of soil frost on growth, composition and respiration of the soil microbial decomposer community. Soil Biol Biochem 43: 2069–2077.
Hendricks DM . (1985) Arizona Soils. In: Haney RA (eds). College of Agriculture, University of Arizona: Tucson, AZ.
Hungate BA, Mau RL, Schwartz E, Caporaso JG, Dijkstra P, van Gestel N et al. (2015). Quantitative microbial ecology through stable isotope probing. Appl and Environ Microb 81: 7570–7581.
Jenkins SN, Rushton SP, Lanyon CV, Whiteley AS, Waite IS, Brookes PC et al. (2010). Taxon-specific responses of soil bacteria to the addition of low level C inputs. Soil Biol Biochem 42: 1624–1631.
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD et al. (2011). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26: 1463–1464.
Kotte O, Zaugg JB, Heinemann M . (2010). Bacterial adaptation through distributed sensing of metabolic fluxes. Mol Syst Biol 6: 355.
Kremling A, Geiselmann J, Ropers D, De Jong H . (2015). Understanding carbon catabolite repression in Escherichia coli using quantitative models. Trends Microbial 23: 99–109.
Kuzyakov Y, Bol R . (2006). Sources and mechanisms of priming effect induced in two grassland soils amended with slurry and sugar. Soil Biol Biochem 38: 747–758.
Kuzyakov Y . (2010). Priming effects: interactions between living and dead organic matter. Soil Biol Biochem 42: 1363–1371.
Lennon JT, Jones SE . (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 9: 119–130.
Letunic I, Bork P . (2011). Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–W478 gkr201.
Liu CM, Aziz M, Kachur S, Hsueh P-R, Huang Y-T, Keim P et al. (2012). BactQuant: an enhanced broad-coverage bacterial quantitative realtime PCR assay. BMC Microbiol 12: 56.
Liu XJA, Sun J, Mau RL, Finley BK, Compson ZG, van Gestel N et al. (2017). Labile carbon input determines the direction and magnitude of the priming effect. Appl Soil Ecol 109: 7–13.
Luo Z, Wang E, Sun OJ . (2016). A meta-analysis of the temporal dynamics of priming soil carbon decomposition by fresh carbon inputs across ecosystems. Soil Biol Biochem 101: 96–103.
Mau RL, Liu CM, Aziz M, Schwartz E, Dijkstra P, Marks JC et al. (2015). Linking soil bacterial biodiversity and soil carbon stability. ISME J 9: 1477–1480.
Mayali X, Weber PK, Mabery S, Pett-Ridge J . (2014). Phylogenetic patterns in the microbial response to resource availability: amino acid incorporation in San Francisco Bay. PloS One 9: e95842.
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618.
Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M et al. (2008). The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform 9: 1.
Mitsui H, Gorlach K, Lee HJ, Hattori R, Hattori T . (2007). Incubation time and media requirements of culturable bacteria from different phylogenetic groups. J Microbiol Meth 30: 103–110.
Moreno-Letelier A, Olemdo G, Eguiarte LE, Martinez-Castilla L, Souza V . (2011). Parallel evolution and horizontal gene transfer of the pst operon in Firmicutes from oligotrophic environments. Int J Evol Biol 2011: 781642.
Morrissey EM, McHugh TA, Preteska L, Hayer M, Dijkstra P, Hungate BA et al. (2015). Dynamics of extracellular DNA decomposition and bacterial community composition in soil. Soil Biol Biochem 86: 42–49.
Morrissey EM, Franklin RB . (2015). Evolutionary history influences the salinity preference of bacterial taxa in wetland soils. Front Microbiol 6: 1013.
Morrissey EM, Mau RL, Schwartz E, Caparosa JG, Dijkstra P, van Gestel N et al. (2016). Phylogenetic organization of bacteria activity. ISME J 10: 2336–2340.
Overkamp W, Ercan O, Herber M, Maris AJ, Kleerebezum M, Kuipers OP . (2015). Physiological and cell morphology adaptation of Bacillus subtilis at near‐zero specific growth rates: a transcriptome analysis. Environ Microbiol 17: 346–363.
Pascault N, Ranjard L, Kaisermann A, Bachar D, Christen R, Terrat S et al. (2013). Stimulation of different functional groups of bacteria by various plant residues as a driver of soil priming effect. Ecosystems 16: 810–822.
Philippot L, Bru D, Saby N, Čuhel J, Arrouays D, Šimek M et al. (2009). Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environ Microbiol 11: 3096–3104.
Phillips RP, Finzi AC, Bernhardt ES . (2011). Enhanced root exudation induces microbial feedbacks to N cycling in a pine forest under long‐term CO2 fumigation. Ecol Lett 14: 187–194.
Poff NL, Olden JD, Vieira NK, Finn DS, Simmons MP, Kondratieff BC . (2006). Functional trait niches of North American lotic insects: traits-based ecological applications in light of phylogenetic relationships. J N Am Benthol Soc 25: 730–755.
Reizer JO, Novotny MJ, Stuiver IN, Saier MH . (1984). Regulation of glycerol uptake by the phosphoenolpyruvate-sugar phosphotransferase system in Bacillus subtilis. J Bacteriol 159: 243–250.
Saier MH . (2014) Mechanisms and Regulation of Carbohydrate Transport in Bacteria. Academic Press Inc.: Orlando, FL, USA.
Sawada K, Funakawa S, Toyota K, Kosaki T . (2015). Potential nitrogen immobilization as influenced by available carbon in Japanese arable and forest soils. Soil Sci Plant Nutr 61: 917–926.
Scharlemann JP, Tanner EV, Hiederer R, Kapos V . (2014). Global soil carbon: understanding and managing the largest terrestrial carbon pool. Carbon Manage 5: 81–91.
Schimel JP, Weintraub MN . (2003). The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: a theoretical model. Soil Biol Biochem 35: 549–563.
Schwartz E . (2007). Characterization of growing microorganisms in soil by stable isotope probing with H218O. Appl Environ Microbiol 73: 2541–2546.
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S et al. (2001). Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67: 4742–4751.
Thomson BC, Ostle NJ, McNamara NP, Oakley S, Whiteley AS, Bailey MJ et al. (2014). Plant soil interactions alter carbon cycling in an upland grassland soil. Front Microbiol 4: 253.
Valvidia-Anistro JA, Eguiarte-Fruns LE, Delgado-Sapién G, Márquez-Zacarías P, Gasca-Pineda J, Learned J et al. (2015). Variability of rRNA operon copy number and growth rate dynamics of Bacillus isolated from an extremely oligotrophic aquatic ecosystem. Front Microbiol 6: 1486.
van Groenigen KJ, Qi X, Osenberg CW, Luo Y, Hungate BA . (2014). Faster decomposition under increased atmospheric CO2 limits soil carbon storage. Science 344: 508–509.
Wang H, Boutton TW, Xu W, Hu G, Jiang P, Bai E . (2015). Quality of fresh organic matter affects priming of soil organic matter and substrate utilization patterns of microbes. Sci Rep 5: 10102.
Webb CO, Ackerly DD, McPeek MA, Donoghue MJ . (2002). Phylogenies and community ecology. Annu Rev Ecol Syst 33: 475–505.
Will C, Thürmer A, Wollherr A, Nacke H, Herold N, Schrumpf M et al. (2010). Horizon-specific bacterial community composition of German grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl Environ Microbiol 76: 6751–6759.
Yin H, Wheeler E, Phillips RP . (2014). Root-induced changes in nutrient cycling in forests depend on exudation rates. Soil Biol Biochem 78: 213–221.
Zhang W, Chao L, Yang Q, Wang Q, Fang Y, Wang S . (2016). Litter quality mediated nitrogen effect on plant litter decomposition regardless of soil fauna presence. Ecology 97: 2834–2843.
Acknowledgements
This research was supported by grants from the National Science Foundation (EAR-1124078 and DEB-1321792) and the Department of Energy's Biological Systems Science Division, Program in Genomic Science.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Morrissey, E., Mau, R., Schwartz, E. et al. Bacterial carbon use plasticity, phylogenetic diversity and the priming of soil organic matter. ISME J 11, 1890–1899 (2017). https://doi.org/10.1038/ismej.2017.43
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2017.43
This article is cited by
-
Relative abundances of bacterial phyla are strong indicators of community-scale microbial growth rates in soil
Environmental Microbiome (2025)
-
Clarifying the role of microbial communities in carbon loss from rhizosphere priming of contrasting crop species under elevated atmospheric CO2
Plant and Soil (2025)
-
Microbial nutrient limitations and chemical composition of soil organic carbon regulate the organic carbon mineralization and temperature sensitivity in forest and grassland soils
Plant and Soil (2025)
-
Adding glucose at a uniform amount or according to soil organic carbon content does not cause significant differences in soil priming effects across a broad grassland transect
Soil Ecology Letters (2025)
-
Distinctive mechanisms of soil priming in different stages and its response to nitrogen addition along a temperate forest elevation gradient
Soil Ecology Letters (2025)