Abstract
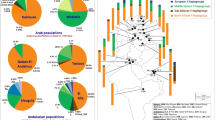
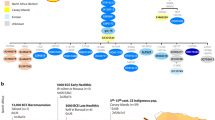
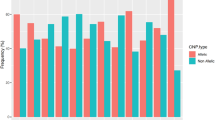
During successive historical periods, Tunisia has been a crossroads of multiple civilizations and their corresponding key population movements. The aim of this study was to provide genetic information relating to the mixed origin of the Tunisian population, and to analyze its genetic relationship with other North African and Mediterranean populations. A set of 16 Alu and 3 Alu/STR compound systems has been analyzed in 268 autochthonous Tunisians from the north-center and the south of the country. Our two sampled populations showed no significant differentiation from one another in any of the three Alu/STR compound systems, whereas the analysis of the 16 Alu markers revealed a significant genetic differentiation between them. A sub-Saharan component shown by the three Alu/STR combinations is more noticeable in our north-center sample than in that of the south. The presence of two Alu/STR combinations specific to North African ancestral populations also suggests that the ancient Berber component is relatively more substantial in the north and center regions than in the south. Our Tunisian samples cluster together with other Berber samples from Morocco and Algeria, underpinning the genetic similarity among North Africans regardless of their current linguistic status (Berber or Arabic).
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Gragueb, A. & Mtimet, A. La Préhistoire en Tunisie et au Maghreb (Tunis Édition ALIF: Tunisia, 1989).
Buresi, P. Géo-histoire de l’islam (Belin-Sup.Histoire: Paris, France, 2005)..
Chaabani, H., Helal, A. N., van Loghem, E., Langaney, A., Benammar Elgaaied, A., Rivat Peran, L. et al. Genetic study of Tunisian Berbers. I. Gm, Am and Km immunoglobulin allotypes and ABO blood groups. J. Immunogenet. 11, 107–113 (1984).
Khodjet-el-khil, H., Fadhlaoui-zid, K., Gusmao, L., Alves, C., Benammar-Elgaaied, A. & Amorim, A. Substructure of a Tunisian Berber population as inferred from 15 autosomal short tandem repeat loci. Hum. Biol. 80, 435–448 (2008).
Ennafaa, H., Amor, M. B., Yacoubi-Loueslati, B., Khodjet el-khil, H., Gonzalez-Perez, E., Moral, P. et al. Alu polymorphisms in Jerba Island population (Tunisia): comparative study in Arab and Berber groups. Ann. Hum. Biol. 33, 634–640 (2006).
Frigi, S., Pereira, F., Pereira, L., Yacoubi, B., Gusmão, L., Alves, C. et al. Data for Y-chromosome haplotypes defined by 17 STRs (AmpFLSTR Yfiler) in two Tunisian Berber communities. Forensic. Sci. Int. 27, 80–83 (2006).
Chaabani, H. & Cox, D. W. Genetic characterisation and origin of Tunisian Berbers. Hum. Hered. 38, 308–316 (1988).
Bahri, R., Esteban, E., Moral, P. & Chaabani, H. New insights into the genetic history of Tunisians: data from Alu insertion and apolipoprotein E gene polymorphisms. Ann. Hum. Biol. 35, 22–33 (2008).
Watkins, W. S., Ricker, C. E., Bamshad, M. J., Carroll, M. L., Nguyen, S. V., Batzer, M. A. et al. Patterns of ancestral human diversity: an analysis of Alu-insertion and restriction-site polymorphisms. Am. J. Hum. Genet. 68, 738–752 (2001).
Flores, C., Maca-Meyer, N., González, A. M. & Cabrera, V. M. Northwest African distribution of the CD4 Alu/microsatellite haplotypes. Ann. Hum. Genet. 64, 321–327 (2000).
González-Pérez, E., Moral, P., Via, M., Vona, G., Varesi, L., Santamaria, J. et al. The ins and outs of population relationships in west-Mediterranean islands: data from autosomal Alu polymorphisms and Alu/STR compound systems. J. Hum. Genet. 52, 999–1010 (2007).
González-Pérez, E., Esteban, E., Via, M., Gaya-Vidal, M., Athanasiadis, G., Dugoujon, J. M. et al. Population relationships in the Mediterranean revealed by autosomal genetic data (Alu and Alu/STR compound systems). Am. J. Phys. Anthrop. 141, 430–439 (2010).
Guo, S. & Thomson, E. Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics 48, 361–372 (1992).
Nei, M. Molecular Evolutionary Genetics (Columbia University Press: New York, 1987).
Raymond, M. & Rousset, F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Heredity 86, 248–249 (1995).
Excoffier, L., Laval, G. & Schneider, S. ARLEQUIN ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinf. Online 1, 47–50 (2005).
Reynolds, J., Weir, B. S. & Cockerman, C. C. Estimation of the coancestry coefficient: basis for a short term genetic distance. Genetics 105, 767–779 (1983).
Wang, J. Maximum-likelihood estimation of admixture proportions from genetic data. Genetics 164, 747–765 (2003).
Tishkoff, S. A., Dietzsch, E., Speed, W., Pakstis, A. J., Kidd, J. R., Cheung, K. et al. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science 271, 1380–1387 (1996).
Plaza, S., Calafell, F., Helal, A., Bouzerna, N., Lefranc, G., Bertranpetit, J. et al. Joining the pillars of Hercules: mtDNA sequences show multidirectional gene flow in the western Mediterranean. Ann. Hum. Genet. 67, 312–328 (2003).
Cruciani, F., La Fratta, R., Trombetta, B., Santolamazza, P., Sellitto, D., Colomb, E B. et al. Tracing past human male movements in northern/eastern Africa and western Eurasia: new clues from Y-chromosomal haplogroups E-M78 and J-M12. Mol. Biol. Evol. 24, 1300–1311 (2007).
Chaabani, H., Bech-Hansen, N. T. & Cox, D. W. Restriction fragment length polymorphisms associated with immunoglobulin heavy chain gamma genes in Tunisians. Hum. Genet. 73, 110–113 (1989).
Turchi, C., Buscemi, L., Giacchino, E., Onofri, V., Fendt, L., Parson, W. et al. Polymorphisms of mtDNA control region in Tunisian and Moroccan populations: an enrichment of forensic mtDNA databases with Northern Africa data. Forensic Sci. Int. Genet. 3, 166–172 (2009).
Arredi, B., Poloni, E. S., Parachini, S., Zerjal, T., Fathallah, D. M., Makfrelouf, M. et al. A predominantly Neolithic origin for Y-chromosome variation in North Africa. Am. J. Hum. Genet. 75, 338–345 (2004).
Pons, A., de Beaulieu, J. L., Guiot, J. & Reille, M. Le pollen remonte le temps climatique. La recherche 213, 518–520 (1991).
Ennafaa, H., Cabrera, V. M., Abu-Amero, K. K., González, A. M., B Amor, M., Bouhaha, R. et al. Mitochondrial DNA haplogroup H structure in North Africa. BMC Genet. 10, 8 (2009).
Cherni, L., Fernandes, V., Pereira, J. B., Costa, M. D., Goios, A., Frigi, S. et al. Post-last glacial maximum expansion from Iberia to North Africa revealed by fine characterization of mtDNA H haplogroup in Tunisia. Am. J. Phys. Anthrop. 139, 253–260 (2009).
Coudray, C., Guitard, E., Kandil, M., Harich, N., Melhaoui, M., Baali, A. et al. Study of GM immunoglobulin allotypic system in Berbers and Arabs from Morocco. Am. J. Hum. Biol. 18, 23–34 (2006).
Mahadevan, M. S., Amemiya, C., Jansen, G., Sabourin, L., Baird, S., Neville, C. E. et al. Structure and genomic sequence of the myotonic dystrophy (DM kinase) gene. Hum. Mol. Genet. 2, 299–304 (1993).
Arcot, S. S., Adamson, A. W., Lamerdin, J. E., Kanagy, B., Deininger, P. L., Carrano, A. V. et al. Alu fossil relics—distribution and insertion polymorphism. Genome Res. 6, 1084–1092 (1996).
Carroll, M. L., Roy-Engel, A. M., Nguyen, S. V., Salem, A. H., Vogel, E., Vincent, B. et al. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J. Mol. Biol. 311, 17–40 (2001).
Arcot, S. S., Adamson, A. W., Risch, G. W., LaFleur, J., Robichaux, M. B., Lamerdin, J. E. et al. High-resolution cartography of recently integrated human chromosome 19-specific Alu fossils. J. Mol. Biol. 281, 843–856 (1998).
Arcot, S. S., Wang, Z., Weber, J. L., Deininger, P. L. & Batzer, M. A. Alu repeats: a source for the genesis of primate microsatellites. Genomics 29, 136–144 (1995).
Batzer, M. A., Arcot, S. S., Phinney, J. W., Alegria-Hartman, M., Kass, D. H., Milligan, S. M. et al. Genetic variation of recent Alu insertions in human populations. J. Mol. Evol. 42, 22–29 (1996).
Edwards, M. C. & Gibbs, R. A. A human dimosphism resulting from loss of an Alu. Genomics 14, 590–597 (1992).
Acknowledgements
We thank all the anonymous people for their participation in the study. This research has been supported in part by the Tunisian Ministry of Higher Education within the research unity 05/UR/09-04, by the Spanish Ministry of Educación y Ciencia Grant CGL2008-03955, the Generalitat de Catalunya 2009SGR1408 grant, and by the Universitat de Barcelona grants of the Oficina de Mobilitat i Programes Internacionals and the Vicerrectorat de Relacions Internacionals i Institucionals.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
El Moncer, W., Esteban, E., Bahri, R. et al. Mixed origin of the current Tunisian population from the analysis of Alu and Alu/STR compound systems. J Hum Genet 55, 827–833 (2010). https://doi.org/10.1038/jhg.2010.120
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2010.120
Keywords
This article is cited by
-
Association between an angiotensin-converting enzyme gene polymorphism and Alzheimer’s disease in a Tunisian population
Annals of General Psychiatry (2017)
-
A novel mutation in the AGXT gene causing primary hyperoxaluria type I: genotype–phenotype correlation
Journal of Genetics (2016)
-
Association Between Haptoglobin 2-2 Genotype and Coronary Artery Disease and Its Severity in a Tunisian Population
Biochemical Genetics (2014)
-
Molecular diagnosis of distal renal tubular acidosis in Tunisian patients: proposed algorithm for Northern Africa populations for the ATP6V1B1, ATP6V0A4 and SCL4A1genes
BMC Medical Genetics (2013)
-
Lack of Association of NOS3 and ACE Gene Polymorphisms with Coronary Artery Disease in Southern Tunisia
Biochemical Genetics (2013)