Abstract
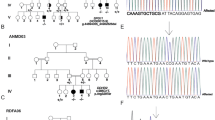
We report an association between a new causative gene and spastic paraplegia, which is a genetically heterogeneous disorder. Clinical phenotyping of one consanguineous family followed by combined homozygosity mapping and whole-exome sequencing analysis. Three patients from the same family shared common features of progressive complicated spastic paraplegia. They shared a single homozygous stretch area on chromosome 6. Whole-exome sequencing revealed a homozygous mutation (c.853_871del19) in the gene coding the kinesin light chain 4 protein (KLC4). Meanwhile, the unaffected parents and two siblings were heterozygous and one sibling was homozygous wild type. The 19 bp deletion in exon 6 generates a stop codon and thus a truncated messenger RNA and protein. The association of a KLC4 mutation with spastic paraplegia identifies a new locus for the disease.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Lo Giudice, T., Lombardi, F., Santorelli, F. M., Kawarai, T. & Orlacchio, A. Hereditary spastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms. Exp. Neurol. 261, 518–539 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
DePristo, M. A., Banks, E., Poplin, R., Garimella, K. V., Maguire, J. R., Hartl, C. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Cingolani, P., Platts, A., Wang le, L., Coon, M., Nguyen, T., Wang, L. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
Teer, J. K., Green, E. D., Mullikin, J. C. & Biesecker, L. G. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics 28, 599–600 (2012).
Gormez, Z., Bakir-Gungor, B. & Sagiroglu, M. S. HomSI: a homozygous stretch identifier from next-generation sequencing data. Bioinformatics 30, 445–447 (2014).
Robinson, J. T., Thorvaldsdottir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Zhu, H., Lee, H. Y., Tong, Y., Hong, B. S., Kim, K. P., Shen, Y. et al. Crystal structures of the tetratricopeptide repeat domains of kinesin light chains: insight into cargo recognition mechanisms. PLoS ONE 7, e33943 (2012).
Endow, S. A., Kull, F. J. & Liu, H. Kinesins at a glance. J. Cell Sci. 123, 3420–3424 (2010).
Kamal, A. & Goldstein, L. S. Principles of cargo attachment to cytoplasmic motor proteins. Curr. Opin. Cell Biol. 14, 63–68 (2002).
Gindhart, J. G. Jr., Desai, C. J., Beushausen, S., Zinn, K. & Goldstein, L. S. Kinesin light chains are essential for axonal transport in Drosophila. J. Cell Biol. 141, 443–454 (1998).
Rahman, A., Kamal, A., Roberts, E. A. & Goldstein, L. S. Defective kinesin heavy chain behavior in mouse kinesin light chain mutants. J. Cell Biol. 146, 1277–1288 (1999).
Acknowledgements
Exome sequencing experiments of this study supported by The Republic of Turkey Ministry of Development Infrastructure Grant (no: 2011K120020) and BILGEM—TUBITAK (The Scientific and Technological Research Council of Turkey) (grant no: T439000)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Bayrakli, F., Poyrazoglu, H., Yuksel, S. et al. Hereditary spastic paraplegia with recessive trait caused by mutation in KLC4 gene. J Hum Genet 60, 763–768 (2015). https://doi.org/10.1038/jhg.2015.109
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2015.109
This article is cited by
-
Analysis of pooled genome sequences from Djallonke and Sahelian sheep of Ghana reveals co-localisation of regions of reduced heterozygosity with candidate genes for disease resistance and adaptation to a tropical environment
BMC Genomics (2019)
-
Kinesins in neurological inherited diseases: a novel motor-domain mutation in KIF5A gene in a patient from Southern Italy affected by hereditary spastic paraplegia
Acta Neurologica Belgica (2018)
-
Hereditary Spastic Paraplegia: Clinical and Genetic Hallmarks
The Cerebellum (2017)