Abstract
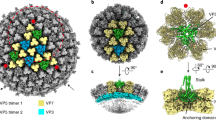
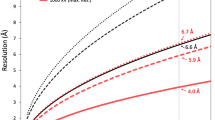
Non-enveloped virus particles (those that lack a lipid-bilayer membrane) must breach the membrane of a target host cell to gain access to its cytoplasm. So far, the molecular mechanism of this membrane penetration step has resisted structural analysis. The spike protein VP4 is a principal component in the entry apparatus of rotavirus, a non-enveloped virus that causes gastroenteritis and kills 440,000 children each year1. Trypsin cleavage of VP4 primes the virus for entry by triggering a rearrangement that rigidifies the VP4 spikes2. We have determined the crystal structure, at 3.2 Å resolution, of the main part of VP4 that projects from the virion. The crystal structure reveals a coiled-coil stabilized trimer. Comparison of this structure with the two-fold clustered VP4 spikes in a ∼12 Å resolution image reconstruction from electron cryomicroscopy of trypsin-primed virions shows that VP4 also undergoes a second rearrangement, in which the oligomer reorganizes and each subunit folds back on itself, translocating a potential membrane-interaction peptide from one end of the spike to the other. This rearrangement resembles the conformational transitions of membrane fusion proteins of enveloped viruses3,4,5,6.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Parashar, U. D., Hummelman, E. G., Bresee, J. S., Miller, M. A. & Glass, R. I. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9, 565–572 (2003)
Crawford, S. E. et al. Trypsin cleavage stabilizes the rotavirus VP4 spike. J. Virol. 75, 6052–6061 (2001)
Ruigrok, R. W. et al. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J. Gen. Virol. 69, 2785–2795 (1988)
Weissenhorn, W. et al. The ectodomain of HIV-1 env subunit gp41 forms a soluble, alpha-helical, rod-like oligomer in the absence of gp120 and the N-terminal fusion peptide. EMBO J. 15, 1507–1514 (1996)
Modis, Y., Ogata, S., Clements, D. & Harrison, S. C. Structure of the dengue virus envelope protein after membrane fusion. Nature 427, 313–319 (2004)
Gibbons, D. L. et al. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427, 320–325 (2004)
Dormitzer, P. R., Sun, Z.-Y. J., Wagner, G. & Harrison, S. C. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21, 885–897 (2002)
Mackow, E. R. et al. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc. Natl Acad. Sci. USA 85, 645–649 (1988)
Dowling, W., Denisova, E., LaMonica, R. & Mackow, E. R. Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J. Virol. 74, 6368–6376 (2000)
Graham, K. L. et al. Integrin-using rotaviruses bind α2β1 integrin α2 I domain via VP4 DGE sequence and recognize αXβ2 and αVβ3 by using VP7 during cell entry. J. Virol. 77, 9969–9978 (2003)
Ruggeri, F. M. & Greenberg, H. B. Antibodies to the trypsin cleavage peptide VP8 neutralize rotavirus by inhibiting binding of virions to target cells in culture. J. Virol. 65, 2211–2219 (1991)
Offit, P. A., Shaw, R. D. & Greenberg, H. B. Passive protection against rotavirus-induced diarrhea by monoclonal antibodies to surface proteins vp3 and vp7. J. Virol. 58, 700–703 (1986)
Dormitzer, P. R., Greenberg, H. B. & Harrison, S. C. Proteolysis of monomeric recombinant rotavirus VP4 yields an oligomeric VP5* core. J. Virol. 75, 7339–7350 (2001)
Arias, C. F., Romero, P., Alvarez, V. & Lopez, S. Trypsin activation pathway of rotavirus infectivity. J. Virol. 70, 5832–5839 (1996)
Gilbert, J. M. & Greenberg, H. B. Cleavage of rhesus rotavirus VP4 after arginine 247 is essential for rotavirus-like particle-induced fusion from without. J. Virol. 72, 5323–5327 (1998)
Tihova, M., Dryden, K. A., Bellamy, A. R., Greenberg, H. B. & Yeager, M. Localization of membrane permeabilization and receptor binding sites on the VP4 hemagglutinin of rotavirus: implications for cell entry. J. Mol. Biol. 314, 985–992 (2001)
Taniguchi, K. et al. Identification of cross-reactive and serotype 2-specific neutralization epitopes on VP3 of human rotavirus. J. Virol. 62, 2421–2426 (1988)
Shaw, A. L. et al. Three-dimensional visualization of the rotavirus hemagglutinin structure. Cell 74, 693–701 (1993)
Yeager, M., Berriman, J. A., Baker, T. S. & Bellamy, A. R. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 13, 1011–1018 (1994)
Liemann, S., Chandran, K., Baker, T. S., Nibert, M. L. & Harrison, S. C. Structure of the reovirus membrane-penetration protein, µ1, in a complex with its protector protein, σ3. Cell 108, 283–295 (2002)
Weeks, C. M. & Miller, R. The design and implementation of SnB v2.0. J. Appl. Crystallogr. 32, 120–124 (1999)
Padilla, J. E. & Yeates, T. O. A statistic for local intensity differences: robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr. D 59, 1124–1130 (2003)
Crowther, R. A. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Phil. Trans. R. Soc. Lond. B 261, 221–230 (1971)
Lawton, J. A. & Prasad, B. V. V. Automated software package for icosahedral virus reconstruction. J. Struct. Biol. 116, 209–215 (1996)
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999)
Wriggers, W., Milligan, R. A. & McCammon, J. A. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 125, 185–195 (1999)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Kraulis, J. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991)
Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from interfacial and thermodynamic properties of hydrocarbons. Prot. Struct. Funct. Genet. 11, 281–296 (1991)
Yeager, M., Dryden, K. A., Olson, N. H., Greenberg, H. B. & Baker, T. S. Three-dimensional structure of rhesus rotavirus by cryoelectron microscopy and image reconstruction. J. Cell Biol. 110, 2133–2144 (1990)
Acknowledgements
We thank M. Babyonyshev for technical assistance; T. Yeates for help in analysing the crystal twinning disorder; H. Greenberg for cloned genes and recombinant baculoviruses; E. Vogan for help with data collection and analysis; and the staff of Advanced Photon Source beamline ID-19 (Argonne National Laboratory) and Cornell High Energy Synchrotron Source beamlines F1 and A1. We acknowledge the use of electron cryomicroscopy facilities at the National Center for Macromolecular Imaging funded by NIH at Baylor College of Medicine. This work was supported by an NIH grant and an Ellison Medical Foundation New Investigator in Global Infectious Diseases award to P.R.D., by an NIH grant to B.V.V.P., and by an NIH grant to S.C.H., who is a Howard Hughes Medical Institute Investigator.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure 1
Comparison of the rotavirus VP4 F'G loop and the alphavirus E1 fusion loop. Includes an amino acid sequence comparison and a structural comparison. (PDF 158 kb)
Supplementary Figure 2
Sample of electron density. An image from an unaveraged simulated annealing omit map is shown. (PDF 1474 kb)
Supplementary Methods
Details of the crystallographic structure determination. Includes a description of the twinning disorder in the crystals. (DOC 35 kb)
Supplementary Table 1
Neutralization escape mutations. Mutations that map to the VP5* fragment are assigned to epitopes based on the VP5CT structure. (DOC 62 kb)
Supplementary Table 2
Rotavirus VP4 sequences used to assess variability and obtain a consensus sequence. The variability in these sequences is mapped onto the surfaces of the VP8* core and VP5* antigen domain in Fig. 3a, c. The consensus sequence is used to assess the similarity between the VP4 F'G loop and the alphavirus E1 fusion loop in Supplementary Fig. 1. (DOC 37 kb)
Supplementary Table 3
Alphavirus E1 sequences used to obtain a consensus. The consensus sequence is used to assess the similarity between the VP4 F'G loop and the alphavirus E1 fusion loop in Supplementary Fig. 1. (DOC 26 kb)
Supplementary Table 4
X-ray diffraction data collection, phasing, and refinement statistics. (DOC 47 kb)
Rights and permissions
About this article
Cite this article
Dormitzer, P., Nason, E., Venkataram Prasad, B. et al. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 430, 1053–1058 (2004). https://doi.org/10.1038/nature02836
Received:
Accepted:
Issue date:
DOI: https://doi.org/10.1038/nature02836
This article is cited by
-
Penetration of non-enveloped viruses
Nature Microbiology (2021)
-
Whole genome and in-silico analyses of G1P[8] rotavirus strains from pre- and post-vaccination periods in Rwanda
Scientific Reports (2020)
-
Porcine rotavirus mainly infects primary porcine enterocytes at the basolateral surface
Veterinary Research (2019)
-
Molecular characterisation of wild-type G1P[8] and G3P[8] rotaviruses isolated in Vietnam 2008 during a vaccine trial
Archives of Virology (2016)
-
Carbohydrate recognition by rotaviruses
Journal of Structural and Functional Genomics (2014)