Abstract
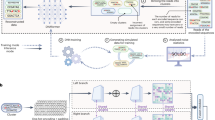
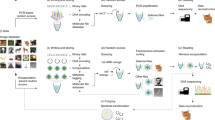
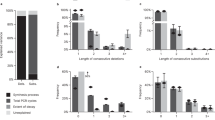
Digital production, transmission and storage have revolutionized how we access and use information but have also made archiving an increasingly complex task that requires active, continuing maintenance of digital media. This challenge has focused some interest on DNA as an attractive target for information storage1 because of its capacity for high-density information encoding, longevity under easily achieved conditions2,3,4 and proven track record as an information bearer. Previous DNA-based information storage approaches have encoded only trivial amounts of information5,6,7 or were not amenable to scaling-up8, and used no robust error-correction and lacked examination of their cost-efficiency for large-scale information archival9. Here we describe a scalable method that can reliably store more information than has been handled before. We encoded computer files totalling 739 kilobytes of hard-disk storage and with an estimated Shannon information10 of 5.2 × 106 bits into a DNA code, synthesized this DNA, sequenced it and reconstructed the original files with 100% accuracy. Theoretical analysis indicates that our DNA-based storage scheme could be scaled far beyond current global information volumes and offers a realistic technology for large-scale, long-term and infrequently accessed digital archiving. In fact, current trends in technological advances are reducing DNA synthesis costs at a pace that should make our scheme cost-effective for sub-50-year archiving within a decade.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
Primary accessions
Sequence Read Archive
Data deposits
Data are available at http://www.ebi.ac.uk/goldman-srv/DNA-storage and in the Sequence Read Archive (SRA) with accession number ERP002040.
References
Baum, E. B. Building an associative memory vastly larger than the brain. Science 268, 583–585 (1995)
Cox, J. P. L. Long-term data storage in DNA. Trends Biotechnol. 19, 247–250 (2001)
Anchordoquy, T. J. & Molina, M. C. Preservation of DNA. Cell Preserv. Technol. 5, 180–188 (2007)
Bonnet, J. et al. Chain and conformation stability of solid-state DNA: implications for room temperature storage. Nucleic Acids Res. 38, 1531–1546 (2010)
Clelland, C. T., Risca, V. & Bancroft, C. Hiding messages in DNA microdots. Nature 399, 533–534 (1999)
Kac, E. Genesis (1999); available at http://www.ekac.org/geninfo.html (accessed, 10 May 2012)
Ailenberg, M. & Rotstein, O. D. An improved Huffman coding method for archiving text, images, and music characters in DNA. Biotechniques 47, 747–754 (2009)
Gibson, D. G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010)
Church, G. M., Gao, Y. & Kosuri, S. Next-generation digital information storage in DNA. Science 337, 1628 (2012)
MacKay, D. J. C. Information Theory, Inference, and Learning Algorithms (Cambridge Univ. Press, 2003)
Erlich, H. A., Gelfand, D. & Sninsky, J. J. Recent advances in the polymerase chain reaction. Science 252, 1643–1651 (1991)
Monaco, A. P. & Larin, Z. YACs, BACs, PACs and MACs: artificial chromosomes as research tools. Trends Biotechnol. 12, 280–286 (1994)
Carr, P. A. & Church, G. M. Genome engineering. Nature Biotechnol. 27, 1151–1162 (2009)
Willerslev, E. et al. Ancient biomolecules from deep ice cores reveal a forested southern Greenland. Science 317, 111–114 (2007)
Green, R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010)
Kari, L. & Mahalingam, K. in Algorithms and Theory of Computation Handbook Vol. 2, 2nd edn (eds Atallah, M. J. & Blanton, M. ) 31-1–31-24 (Chapman & Hall, 2009)
Păun, G., Rozenberg, G. & Salomaa, A. DNA Computing: New Computing Paradigms (Springer, 1998)
Watson, J. D. & Crick, F. H. C. Molecular structure of nucleic acids. Nature 171, 737–738 (1953)
Niedringhaus, T. P., Milanova, D., Kerby, M. B., Snyder, M. P. & Barron, A. E. Landscape of next-generation sequencing technologies. Anal. Chem. 83, 4327–4341 (2011)
LeProust, E. M. et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 38, 2522–2540 (2010)
Massingham, T. & Goldman, N. All Your Base: a fast and accurate probabilistic approach to base calling. Genome Biol. 13, R13 (2012)
Gantz, J. & Reinsel, D. Extracting Value from Chaos (IDC, 2011)
Brand, S. The Clock of the Long Now (Basic Books, 1999)
Digital. archiving. History flushed. Economist 403, 56–57 (28 April 2012); available at http://www.economist.com/node/21553410 (2012)
Bessone, N., Cancio, G., Murray, S. & Taurelli, G. Increasing the efficiency of tape-based storage backends. J. Phys. Conf. Ser. 219, 062038 (2010)
Baker, M. et al. in Proc. 1st ACM SIGOPS/EuroSys European Conf. on Computer Systems (eds Berbers, Y. & Zwaenepoel, W. ) 221–234 (ACM, 2006)
Yuille, M. et al. The UK DNA banking network: a “fair access” biobank. Cell Tissue Bank. 11, 241–251 (2010)
Global Crop Diversity Trust Svalbard Global Seed Vault. (2012); available at http://www.croptrust.org/main/content/svalbard-global-seed-vault (accessed, 10 May 2012)
Acknowledgements
At the University of Cambridge: D. MacKay and G. Mitchison for advice on codes for run-length-limited channels. At CERN: B. Jones for discussions on data archival. At EBI: A. Löytynoja for custom multiple sequence alignment software, H. Marsden for computing base calls and for detecting an error in the original parity-check encoding, T. Massingham for computing base calls and advice on code theory and K. Gori, D. Henk, R. Loos, S. Parks and R. Schwarz for assistance with revisions to the manuscript. In the Genomics Core Facility at EMBL Heidelberg: V. Benes for advice on Next-Generation Sequencing protocols, D. Pavlinić for sequencing and J. Blake for data handling. C.D. is supported by a fellowship from the Swiss National Science Foundation (grant 136461). B.S. is supported by an EMBL Interdisciplinary Postdoctoral Fellowship under Marie Curie Actions (COFUND).
Author information
Authors and Affiliations
Contributions
N.G. and E.B. conceived and planned the project and devised the information-encoding methods. P.B. advised on oligo design and Next-Generation Sequencing protocols, prepared the DNA library and managed the sequencing process. S.C. and E.M.L. provided custom oligonucleotides. N.G. wrote the software for encoding and decoding information into/from DNA and analysed the data. N.G., E.B., C.D. and B.S. modelled the scaling properties of DNA storage. N.G. wrote the paper with discussions and contributions from all other authors. N.G. and C.D. produced the figures.
Corresponding author
Ethics declarations
Competing interests
S.C. and E.M.L. are employees of Agilent Technologies, a commercial provider of OLS pools. N.G. and E.B. are named inventors on a patent application on technologies described in this work.
Supplementary information
Supplementary Information 1
This file contains Supplementary Tables 1-4, Supplementary Figures 1-9, Supplementary Methods and Data, a Supplementary Discussion and Supplementary references. This file was replaced on 14 February 2013 to correct the DNA sequence in Supplementary Figure 8, which was misaligned. (PDF 2027 kb)
Supplementary Information 2
This file contains the full formal specification of the digital information encoding scheme. (PDF 244 kb)
Supplementary Information 3
This file contains FastQC QC report on Illumina HiSeq 2000 sequencing run. (PDF 411 kb)
Supplementary Data 1
This zipped file contains the five original files encoded and decoded in this study, namely wssnt10.txt (ASCII text file containing text of all 154 Shakespeare sonnets), watsoncrick.pdf (PDF of Watson & Crick’s (1953) paper describing the structure of DNA), MLK_excerpt_VBR_45-85.mp3 (MP3 file containing a 26 s excerpt from Martin Luther King's 1963 "I Have A Dream" speech), EBI.jp2 (JPEG 2000 format medium resolution colour photograph of the European Bioinformatics Institute) and View_huff3.cd.new (ASCII text file defining the Huffman code used to convert bytes of encoded files to base 3). (ZIP 646 kb)
Supplementary Data 2
This file contains the GATK ErrorRatePerCycle report on Illumina HiSeq 2000 sequencing run. (TXT 6 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Goldman, N., Bertone, P., Chen, S. et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 494, 77–80 (2013). https://doi.org/10.1038/nature11875
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/nature11875
This article is cited by
-
Magnetic DNA random access memory with nanopore readouts and exponentially-scaled combinatorial addressing
Scientific Reports (2023)
-
Performance analysis of DNA crossbar arrays for high-density memory storage applications
Scientific Reports (2023)
-
Digital data storage on DNA tape using CRISPR base editors
Nature Communications (2023)
-
Towards high-density storage of text and images into DNA by the “Xiao-Pang” codec system
Science China Life Sciences (2023)
-
A Novel Image Encryption Scheme for DNA Storage Systems Based on DNA Hybridization and Gene Mutation
Interdisciplinary Sciences: Computational Life Sciences (2023)

Thomas Dandekar
Light-gated DNA storage is essential
COMMENT ON N. Goldman, P. Bertone, S. Chen, C. Dessimoz, E. LeProust, B. Sipos & E. Birney Nature 494, 77-80 (2013)
Goldman et al. (1) show that DNA can in fact be used as solid and permanent data storage.
In a certain sense this is obvious (we all use it as genetic storage not only all our life, but in fact all our evolutionary history long). However, demonstrating the ?obvious?, Goldman et al. (1) transformed the general statement into a strong proof-of-concept example for the potential of DNA as a storage technology. Arguably, the key steps presented were already shown recently (2) or, at least conceptual, even long before (3,4). Nevertheless, the strong proof of concept shown by Goldman et al.1 in their inspiring paper is a milestone towards using DNA information storage technology: They describe a scalable method to reliably store large volumes of information in DNA with 100% accuracy for large-scale, long-term and infrequently accessed digital archiving.
However, we argue here that we need a critical technology advance more before a junction between electronic data processing and molecular data storage and processing can really take off. There is a serious threat that otherwise DNA storage will never get momentum: This risk is supported for instance by the up-till now failure (in spite of inspiring inventions) to really translate DNA computing (5) and RNA logical gates (6) into a technology delivering not only ?interesting? results but technological power and spread.
To achieve a robust, addressable and user-friendly molecular information processing technology we argue that a combination of the advantages of nanotechnology, molecular biology and external, user-specific control is necessary, similarly as PCR took off combining primer directed specific recall of information with robust, heat stable Taq polymerase. We claim that a direct feedback of technical input into molecular circuits is necessary as well as direct feed-into of the molecular result into technical processing machinery for DNA storage to take off (e.g. not requiring a technical apparatus and cumbersome sequencing steps to decipher the stored information).
A direct connection from molecular processing in cells and DNA to technical computers is necessary to achieve speed and calculation potential. Electronic properties of DNA (7) are difficult to handle. We suggest for linking DNA information processing to in silico processing step-by-step in an efficient way light-gated proteins (8). Light-gated proteins allow (i) control of their own and other enzyme activities, (ii) gene expression and protein-protein interactions, as well as (iii) to achieve patterning and directing cell to cell communication and integration of circuits. Containment features control the high biological repair and replication potential of such biobricks (9) which together achieve extremely robust active DNA storage technology without negative side-effects or uncontrolled risks.
Critical steps needed to be achieved and a blueprint of the design of the active DNA storage we currently explore include light gated protein constructs to achieve rapid light-directed DNA synthesis as well as direct DNA-sequence readout via optical signals.
In conclusion, our claim to the recent work by Goldman et al. (1) is that active DNA storage technology is critical so that DNA storage can really take off and will be broadly used. This includes user directed molecular DNA synthesis and sequencing, in particular by light-gated proteins. Without active DNA storage, the technology will remain a technological tour de force, in ten years maybe cheap but slow in effective information recall, let alone calculations.
					
Thomas Dandekar [1,2], Daniel Lopez<sup class="footnote">3</sup>, Dominik Schaack <sup class="footnote">1</sup>
1 -Dept. of Bioinformatics, Biocenter, University of Würzburg, Am Hubland, 97074 Würzburg, Germany. e-mail: dandekar@biozentrum.uni-wuerzburg.de; phone ++49-931-318-4551; Fax -4552;
2 -EMBL, Meyerhofstrasse 1, 69117 Heidelberg, Germany
3 -Research center for infectious disease, Josef Schneider Str. 2/ D15, 97080 Würzburg, Germany