Abstract
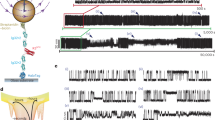
Free-energy-landscape formalisms provide the fundamental conceptual framework for physical descriptions of how proteins and nucleic acids fold into specific three-dimensional structures1,2. Although folding landscapes are difficult to measure experimentally, recent theoretical work by Hummer and Szabo3 has shown that landscape profiles can be reconstructed from non-equilibrium single-molecule force spectroscopy measurements using an extension of the Jarzynski equality4. This method has been applied to simulations5,6 and experiments7,8 but never validated experimentally. We tested it using force–extension measurements on DNA hairpins with distinct, sequence-dependent folding landscapes. Quantitative agreement was found between the landscape profiles obtained from the non-equilibrium reconstruction and those from equilibrium probability distributions9. We also tested the method on a riboswitch aptamer with three partially folded intermediate states, successfully reconstructing the landscape but finding some states difficult to resolve owing to low occupancy or overlap of the potential wells. These measurements validate the landscape-reconstruction method and provide a new test of non-equilibrium work relations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Onuchic, J. N., Luthey-Schulten, Z. & Wolynes, P. G. Theory of protein folding: The energy landscape perspective. Annu. Rev. Phys. Chem. 48, 545–600 (1997).
Bryngelson, J. D. & Wolynes, P. G. Spin glasses and the statistical mechanics of protein folding. Proc. Natl Acad. Sci. USA 84, 7524–7528 (1987).
Hummer, G. & Szabo, A. Free energy reconstruction from nonequilibrium single-molecule pulling experiments. Proc. Natl Acad. Sci. USA 98, 3658–3661 (2001).
Jarzynski, C. Nonequilibrium equality for free energy differences. Phys. Rev. Lett. 78, 2690 (1997).
Park, S. & Schulten, K. Calculating potentials of mean force from steered molecular dynamics simulations. J. Chem. Phys. 120, 5946–5961 (2004).
Minh, D. D. L. Free-energy reconstruction from experiments performed under different biasing programs. Phys. Rev. E 74, 061120 (2006).
Liphardt, J., Dumont, S., Smith, S. B., Tinoco, I. J. & Bustamante, C. Equilibrium information from nonequilibrium measurements in an experimental test of Jarzynski’s equality. Science 296, 1832–1835 (2002).
Harris, N. C., Song, Y. & Kiang, C-H. Experimental free energy surface reconstruction from single-molecule force spectroscopy using Jarzynski’s equality. Phys. Rev. Lett. 99, 068101 (2007).
Woodside, M. T. et al. Direct measurement of the full sequence-dependent folding landscape of a nucleic acid. Science 314, 1001–1004 (2006).
Petrey, D. & Honig, B. Protein structure prediction: Inroads to biology. Mol. Cell. 20, 811–819 (2005).
Bradley, P., Misura, K. M. S. & Baker, D. Toward high-resolution de novo structure prediction for small proteins. Science 309, 1868–1871 (2005).
Fersht, A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding (Freeman, 1999).
Dobson, C. M. Protein folding and misfolding. Nature 426, 884–890 (2003).
Hyeon, C. & Thirumalai, D. Can energy landscape roughness of proteins and RNA be measured by using mechanical unfolding experiments? Proc. Natl Acad. Sci. USA 100, 10249–10253 (2003).
Kim, P. S. & Baldwin, R. L. Intermediates in the folding reactions of small proteins. Annu. Rev. Biochem. 59, 631–660 (1990).
Daggett, V. & Fersht, A. R. Is there a unifying mechanism for protein folding? Trends Biochem. Sci. 28, 18–25 (2003).
Buchner, J. & Kiefhaber, T. Protein Folding Handbook (Wiley-VCH, 2005).
Woodside, M. T., García-García, C. & Block, S. M. Folding and unfoldingsingle RNA molecules under tension. Curr. Opin. Chem. Biol. 12, 640–646 (2008).
Lang, M. J., Asbury, C. L., Shaevitz, J. W. & Block, S. M. An automated two-dimensional optical force clamp for single molecule studies. Biophys. J. 83, 491–501 (2002).
Greenleaf, W. J., Woodside, M. T., Abbondanzieri, E. A. & Block, S. M. Passive all-optical force clamp for high-resolution laser trapping. Phys. Rev. Lett. 95, 208102 (2005).
Gebhardt, J. C. M., Bornschlögl, T. & Rief, M. Full distance-resolved folding energy landscape of one single protein molecule. Proc. Natl Acad. Sci. USA 107, 2013–2018 (2010).
Crooks, G. E. Entropy production fluctuation theorem and thenonequilibrium work relation for free energy differences. Phys. Rev. E 60, 2721 (1999).
Collin, D. et al. Verification of the Crooks fluctuation theorem and recovery of RNA folding free energies. Nature 437, 231–234 (2005).
Ritort, F. Nonequilibrium fluctuations in small systems: From physics to biology. Adv. Chem. Phys. 137, 31–123 (2008).
Woodside, M. T. et al. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl Acad. Sci. USA 103, 6190–6195 (2006).
Smith, S. B., Cui, Y. & Bustamante, C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science 271, 795–798 (1996).
Liphardt, J., Onoa, B., Smith, S. B., Tinoco, I. J. & Bustamante, C. Reversible unfolding of single RNA molecules by mechanical force. Science 292, 733–737 (2001).
Wang, M. D., Yin, H., Landick, R., Gelles, J. & Block, S. M. Stretching DNA with optical tweezers. J. Biophys. 72, 1335–1346 (1997).
Mossa, A., Lorenzo, S. d., Huguet, J. M. & Ritort, F. Measurement of work in single-molecule pulling experiments. J. Chem. Phys. 130, 234116 (2009).
Greenleaf, W. J., Frieda, K. L., Foster, D. A. N., Woodside, M. T. & Block, S. M. Direct observation of hierarchical folding in single riboswitch aptamers. Science 319, 630–633 (2008).
Acknowledgements
We thank A. Szabo, G. Hummer and D. Minh for discussion and comments. This work was supported by the National Institute for Nanotechnology, Canadian Institutes of Health Research grant reference number NHG 91374, PrioNet Canada and the nanoWorks program of Alberta Innovates Technology Solutions.
Author information
Authors and Affiliations
Contributions
M.T.W. designed the experiment. F.W. contributed materials. K.N., H.Y. and M.T.W. made the measurements. A.N.G., A.V. and K.N. analysed the data. A.N.G., A.V. and M.T.W. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 262 kb)
Rights and permissions
About this article
Cite this article
Gupta, A., Vincent, A., Neupane, K. et al. Experimental validation of free-energy-landscape reconstruction from non-equilibrium single-molecule force spectroscopy measurements. Nature Phys 7, 631–634 (2011). https://doi.org/10.1038/nphys2022
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/nphys2022
This article is cited by
-
Mapping the conformational energy landscape of Abl kinase using ClyA nanopore tweezers
Nature Communications (2022)
-
Experimental evidence of symmetry breaking of transition-path times
Nature Communications (2019)
-
Equilibrium free energies from non-equilibrium trajectories with relaxation fluctuation spectroscopy
Nature Physics (2018)
-
Stochastic analysis of time series for the spatial positions of particles trapped in optical tweezers
Scientific Reports (2017)
-
Improved approximations for some polymer extension models
Rheologica Acta (2017)