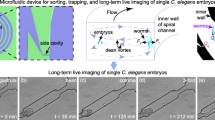
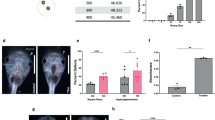
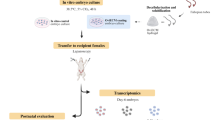
Abstract
Cell culture is an invaluable tool for investigation of basic biological processes. However, technical hurdles including low cell yield, poor cell differentiation and poor attachment to the growth substrate have limited the use of this tool for studies of the genetic model organism Caenorhabditis elegans. This protocol describes a method for the large-scale culture of C. elegans embryo cells. We also describe methods for in vitro RNA interference, fluorescence-activated cell sorting of embryo cells and imaging of cultured cells for patch-clamp electrophysiology studies. Developing embryos are isolated from gravid adult worms. After eggshell removal by enzymatic digestion, embryo cells are dissociated and plated onto glass substrates. Isolated cells terminally differentiate within 24 h. Analysis of gene expression patterns and cell-type frequency suggests that in vitro embryo cell cultures recapitulate the developmental characteristics of L1 larvae. Cultured embryo cells are well suited for physiological analysis as well as molecular and cell biological studies. The embryo cell isolation protocol can be completed in 5–6 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Barr, M.M. Super models. Physiol. Genomics 13, 15–24 (2003).
Strange, K. From genes to integrative physiology: ion channel and transporter biology in Caenorhabditis elegans . Physiol. Rev. 83, 377–415 (2003).
Bloom, L. Genetic and molecular analysis of genes required for axon outgrowth in Caenorhabditis elegans 1–412 (Massachusetts Institute of Technology, Boston, MA, 1993).
Buechner, M., Hall, D.H., Bhatt, H. & Hedgecock, E.M. Cystic canal mutants in Caenorhabditis elegans are defective in the apical membrane domain of the renal (excretory) cell. Dev. Biol. 214, 227–241 (1999).
Christensen, M. & Strange, K. Developmental regulation of a novel outwardly rectifying mechanosensitive anion channel in Caenorhabditis elegans . J. Biol. Chem. 276, 45024–45030 (2001).
Christensen, M. et al. A primary culture system for functional analysis of C. elegans neurons and muscle cells. Neuron 33, 503–514 (2002).
Grishok, A. RNAi mechanisms in Caenorhabditis elegans . FEBS Lett. 579, 5932–5939 (2005).
Kamath, R.S. & Ahringer, J. Genome-wide RNAi screening in Caenorhabditis elegans . Methods 30, 313–321 (2003).
Zhang, Y. et al. Identification of genes expressed in C. elegans touch receptor neurons. Nature 418, 331–335 (2002).
The Axon CNS Guide, A Laboratory Guide to Electrophysiology and Biophysics (Molecular Devices Corporation, Union City, CA, 2006).
Estevez, A.Y., Roberts, R.K. & Strange, K. Identification of store-independent and store-operated Ca2+ conductances in Caenorhabditis elegans intestinal epithelial cells. J. Gen. Physiol. 122, 207–223 (2003).
Yuan, A. et al. The sodium-activated potassium channel is encoded by a member of the Slo gene family. Neuron 37, 765–773 (2003).
Carvelli, L., McDonald, P.W., Blakely, R.D. & DeFelice, L.J. Dopamine transporters depolarize neurons by a channel mechanism. Proc. Natl. Acad. Sci. USA 101, 16046–16051 (2004).
Park, K.H., Hernandez, L., Cai, S.Q., Wang, Y. & Sesti, F. A family of K+ channel ancillary subunits regulate taste sensitivity in Caenorhabditis elegans . J. Biol. Chem. 280, 21893–21899 (2005).
Estevez, A.Y. & Strange, K. Calcium feedback mechanisms regulate oscillatory activity of a TRP-like Ca2+ conductance in C. elegans intestinal cells. J. Physiol. 567, 239–251 (2005).
Bianchi, L. et al. The neurotoxic MEC-4(d) DEG/ENaC sodium channel conducts calcium: implications for necrosis initiation. Nat. Neurosci. 7, 1337–1344 (2004).
Suzuki, H. et al. In vivo imaging of C. elegans mechanosensory neurons demonstrates a specific role for the MEC-4 channel in the process of gentle touch sensation. Neuron 39, 1005–1017 (2003).
Teramoto, T., Lambie, E.J. & Iwasaki, K. Differential regulation of TRPM channels governs electrolyte homeostasis in the C. elegans intestine. Cell Metab. 1, 343–354 (2005).
Mullen, G.P. et al. The Caenorhabditis elegans snf-11 gene encodes a sodium-dependent GABA transporter required for clearance of synaptic GABA. Mol. Biol. Cell 17, 3021–3030 (2006).
Goodman, M.B. & Schwarz, E.M. Transducing touch in Caenorhabditis elegans . Annu. Rev. Physiol. 65, 429–452 (2003).
O'Hagan, R., Chalfie, M. & Goodman, M.B. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat. Neurosci. 8, 43–50 (2005).
Hamill, O.P. & Martinac, B. Molecular basis of mechanotransduction in living cells. Physiol. Rev. 81, 685–740 (2001).
Frokjaer-Jensen, C. et al. Effects of voltage-gated calcium channel subunit genes on calcium influx in cultured C. elegans mechanosensory neurons. J. Neurobiol. 66, 1125–1139 (2006).
Suzuki, H. et al. In vivo imaging of C. elegans mechanosensory neurons demonstrates a specific role for the MEC-4 channel in the process of gentle touch sensation. Neuron 39, 1005–1017 (2003).
Dal Santo, P., Logan, M.A., Chisholm, A.D. & Jorgensen, E.M. The inositol trisphosphate receptor regulates a 50-second behavioral rhythm in C. elegans . Cell 98, 757–767 (1999).
Espelt, M.V., Estevez, A.Y., Yin, X. & Strange, K. Oscillatory Ca2+ signaling in the isolated Caenorhabditis elegans intestine: role of the inositol-1,4,5-trisphosphate receptor and phospholipases C β and γ. J. Gen. Physiol. 126, 379–392 (2005).
Teramoto, T. & Iwasaki, K. Intestinal calcium waves coordinate a behavioral motor program in C. elegans . Cell Calcium 40, 319–327 (2006).
Yan, X. et al. Function of a STIM1 homologue in C. elegans: evidence that store-operated Ca2+ entry is not essential for oscillatory Ca2+ signaling and ER Ca2+ homeostasis. J. Gen. Physiol. 128, 459 (2006).
Lorin-Nebel, C., Xing, J., Yan, X. & Strange, K. CRAC channel activity in C. elegans is mediated by Orai1 and STIM1 homologs and is essential for ovulation and fertility. J. Physiol. 580, 67–85 (2007).
Cinar, H., Keles, S. & Jin, Y. Expression profiling of GABAergic motor neurons in Caenorhabditis elegans . Curr. Biol. 15, 340–346 (2005).
Fox, R.M. et al. A gene expression fingerprint of C. elegans embryonic motor neurons. BMC Genomics 6, 42 (2005).
Colosimo, M.E. et al. Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Curr. Biol. 14, 2245–2251 (2004).
Fukushige, T., Hawkins, M.G. & McGhee, J.D. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198, 286–302 (1998).
Acknowledgements
This work was supported by NIH R01 grants DK51610, DK61168 and GM74229 to K.S.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Strange, K., Christensen, M. & Morrison, R. Primary culture of Caenorhabditis elegans developing embryo cells for electrophysiological, cell biological and molecular studies. Nat Protoc 2, 1003–1012 (2007). https://doi.org/10.1038/nprot.2007.143
Published:
Issue date:
DOI: https://doi.org/10.1038/nprot.2007.143
This article is cited by
-
Effects of Zataria multiflora essential oil on the germinative cells of Echinococcus granulosus
Parasites & Vectors (2021)
-
Polymodal Functionality of C. elegans OLL Neurons in Mechanosensation and Thermosensation
Neuroscience Bulletin (2021)
-
Single-cell RNA profiling links ncRNAs to spatiotemporal gene expression during C. elegans embryogenesis
Scientific Reports (2020)
-
Toxic Effects of Size-tunable Gold Nanoparticles on Caenorhabditis elegans Development and Gene Regulation
Scientific Reports (2018)
-
Developmentally programmed germ cell remodelling by endodermal cell cannibalism
Nature Cell Biology (2016)