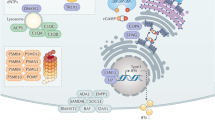
Key Points
-
At least two forms of cell-autonomous immunity operate in higher organisms such as vertebrates: constitutive and inducible. The interferon (IFN) family of cytokines stimulates the inducible gene programme for mobilizing effector functions inside individual host cells.
-
IFN-induced effector proteins operate against most pathogen classes, especially bacteria, protozoan parasites and viruses.
-
Individual bacteria, protozoa and viruses occupy only a tiny fraction of the interior volume of a vertebrate cell. Hence, many IFN-inducible proteins are directly targeted to the site of microbial replication or generate toxic products capable of diffusing large intracellular distances to reach these microorganisms.
-
IFN-induced proteins inhibit intracellular bacteria and protozoa through a variety of mechanisms. These include: oxidative and nitrosative damage caused by cytotoxic gases (generated via IFN-inducible oxidoreductases); the recruitment of the autophagic machinery to deliver microorganisms to lysosomes (by IFN-inducible GTPases and cytosolic receptors); and the depletion of essential amino acids and divalent cations needed for microbial growth (by IFN-induced catabolic enzymes and efflux pumps, respectively).
-
IFN-induced antiviral mechanisms operate across most nucleated cells and at all stages of the viral life cycle, including entry, replication, capsid assembly and release. Several new proteins have recently been discovered that fulfil these different functions.
-
Much scientific effort over the last two decades has focused on how the innate immune system recognizes microbial pathogens. Attention is now beginning to turn towards understanding the effector mechanisms needed to sterilize these infections.
Abstract
Interferons (IFNs) induce the expression of hundreds of genes as part of an elaborate antimicrobial programme designed to combat infection in all nucleated cells — a process termed cell-autonomous immunity. As described in this Review, recent genomic and subgenomic analyses have begun to assign functional properties to novel IFN-inducible effector proteins that restrict bacteria, protozoa and viruses in different subcellular compartments and at different stages of the pathogen life cycle. Several newly described host defence factors also participate in canonical oxidative and autophagic pathways by spatially coordinating their activities to enhance microbial killing. Together, these IFN-induced effector networks help to confer vertebrate host resistance to a vast and complex microbial world.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Marathe, R., Guan, Z., Anandalakshmi, R., Zhao, H. & Dinesh-Kumar, S. P. Study of Arabidopsis thaliana resistome in response to cucumber mosaic virus infection using whole genome microarray. Plant Mol. Biol. 55, 501–520 (2004).
Beutler, B. et al. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu. Rev. Immunol. 24, 353–389 (2006).
Mascie-Taylor, C. G. & Karim, E. The burden of chronic disease. Science 302, 1921–1922 (2003).
Bezbradica, J. J. & Medzhitov, R. Integration of cytokine and heterologous receptor signaling pathways. Nature Immunol. 10, 333–339 (2009).
Kawai, T. & Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650 (2011).
Nathan, C. & Shiloh, M. U. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl Acad. Sci. USA 97, 8841–8848 (2000).
Sadler, A. & Williams, B. R. G. Interferon-inducible antiviral effectors. Nature Rev. Immunol. 8, 559–568 (2008).
Flannagan, R. S., Cosío, G. & Grinstein, S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nature Rev. Microbiol. 7, 355–366 (2009).
Nathan, C. F., Murray, H. W., Weibe, M. E. & Rubin, B. Y. Identification of IFN-γ as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 158, 670–689 (1983). This paper was the first to describe IFNγ as the long sought after macrophage-activating factor required for defence against intracellular pathogens (see also the historical note in reference 11).
MacMicking, J. D. Macrophage activation and host defense. Cell Host Microbe 5, 1–3 (2009).
Hertzog, P., Forster, S. & Samarajiwa, S. Systems biology of interferon responses. J. Interferon Cytokine Res. 31, 5–11 (2011).
Brass, A. L. et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 (2009).
Kumar, D. et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell 140, 731–743 (2010). References 13 and 14 were among the first to use genome-wide small interfering RNA screening against viruses and bacteria to identify new type I and II IFN effectors.
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011). In this study, a comprehensive gain-of-function screen discovered new type I IFN antiviral factors against several major human pathogens, including hepatitis C virus.
Kim, B. H. et al. A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science 332, 717–721 (2011). In this study, family-wide loss-of-function screens and gene-targeted mice were used to identify the 65 kDa GBPs as new IFNγ-inducible proteins that combat intracellular bacterial infection.
Krombach, F. et al. Cell size of alveolar macrophages: an interspecies comparison. Environ. Health Perspect. 105 (Suppl. 5), 1261–1263 (1997).
MacMicking, J. D. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 25, 601–609 (2004).
Martens, S. & Howard, J. The IFN-inducible GTPases. Annu. Rev. Immunol. 22, 558–598 (2006).
Kumar, Y. & Valdivia, R. H. Leading a sheltered life: intracellular pathogens and maintenance of vacuolar compartments. Cell Host Microbe 5, 593–601 (2009).
Tiwari, S., Choi, H. P., Matsuzawa, T., Pypaert, M. & MacMicking, J. D. Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P2 and PtdIns(3,4, 5)P3 promotes immunity to mycobacteria. Nature Immunol. 10, 907–917 (2009).
MacMicking, J. D., Taylor, G. A. & McKinney, J. D. Immune control of tuberculosis by IFN-γ-inducible LRG-47. Science 302, 654–659 (2003). The first report of an IFN-inducible GTPase regulating membrane traffic to compartmentalized pathogens for cell-autonomous defence.
Singh, S. B., Davis, A. S., Taylor, G. A. & Deretic, V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313, 1438–1441 (2006).
Thurston, T. L., Ryzhakov, G., Bloor, S., von Muhlinen, N. & Randow, F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nature Immunol. 10, 1215–1221 (2009).
Thurston, T. L., Wandel, M. P., von Muhlinen, N., Foeglein, A. & Randow, F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418 (2012). References 24 and 25 identify IFN-induced ubiquitin- and glycan-recognition receptors, respectively, that help to bridge the detection of cytosolic bacteria with sequestration by the autophagy pathway.
Nathan, C. & Ding, A. SnapShot: reactive oxygen intermediates (ROI). Cell 140, 952–952. e2 (2010).
Karupiah, G. et al. Inhibition of viral replication by interferon-γ-induced nitric oxide synthase. Science 261, 1445–1448 (1993).
Lambeth, J. D. NOX enzymes and the biology of reactive oxygen. Nature Rev. Immunol. 4, 181–189 (2004).
Fields, K. A. & Hackstadt, T. The chlamydial inclusion: escape from the endocytic pathway. Annu. Rev. Cell Dev. Biol. 18, 221–245 (2002).
Oliveira Gde, A., Lieberman, J. & Barillas-Mury, C. Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science 335, 856–859 (2012).
Sonoda, J. et al. Nuclear receptor ERRα and coactivator PGC-1β are effectors of IFN-γ-induced host defense. Genes Dev. 21, 1909–1920 (2007).
West, A. P. et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472, 476–480 (2011).
Bustamente, J. et al. Germline CYBB mutations that selectively affect macrophages in kindreds with X-linked predisposition to tuberculous mycobacterial disease. Nature Immunol. 12, 213–221 (2011).
Gardet, A. et al. LRRK2 is involved in the IFN-γ response and host response to pathogens. J. Immunol. 185, 5577–5585 (2010).
Moskwa, P. et al. A novel host defense system of airways is defective in cystic fibrosis. Am. J. Respir. Crit. Care Med. 175, 174–183 (2007).
Botteauxa, A., Hosteb, C., Dumontb, J. E., Van Sandeb, J. & Allaouia, A. Potential role of Noxes in the protection of mucosae: H2O2 as a bacterial chemorepellant. Microbes Infect. 11, 537–544 (2009).
Gattas, M. V. et al. Oxidative epithelial host defense is regulated by infectious and inflammatory stimuli. Free Radic. Biol. Med. 47, 1450–1458 (2009).
Flores, M. V. et al. Dual oxidase in the intestinal epithelium of zebrafish larvae has antibacterial properties. Biochem. Biophys. Res. Commun. 400, 164–168 (2010).
MacMicking, J. D. et al. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell 81, 641–650 (1995).
Pollock, J. D. et al. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nature Genet. 9, 202–209 (1995). References 39 and 40 describe the first gene-targeted mice deficient for IFN-inducible effector proteins.
Shiloh, M. U. et al. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10, 29–38 (1999).
Ng, V. H., Cox, J. S., Sousa, A. O., MacMicking, J. D. & McKinney, J. D. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol. Microbiol. 52, 1291–1302 (2004).
Singh, R. et al. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science 322, 1392–1395 (2008).
Myers, J. T., Tsang, A. W. & Swanson, J. A. Localized reactive oxygen and nitrogen intermediates inhibit escape of Listeria monocytogenes from vacuoles in activated macrophages. J. Immunol. 171, 5447–5453 (2003).
Bagshaw, R. D., Mahuran, D. J. & Callahan, J. W. A proteomic analysis of lysosomal integral membrane proteins reveals the diverse composition of this organelle. Mol. Cell. Proteomics 4, 133–143 (2005).
Trost, M. et al. The phagosomal proteome in interferon-γ-activated macrophages. Immunity 30, 143–154 (2009). A comprehensive proteomic study that identified IFN-inducible proteins that are recruited to the phagosome.
Wild, P. et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 (2011).
Mostowy, S. et al. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J. Biol. Chem. 286, 26987–26995 (2011).
Cemma, M., Kim, P. K. & Brumell, J. H. The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy 7, 341–345 (2011).
Yoshikawa, Y. et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nature Cell Biol. 11, 1233–1240 (2009).
Shenoy, A. R. et al. Emerging themes in IFN-γ-induced macrophage immunity by the p47 and p65 GTPase families. Immunobiology 212, 771–784 (2008).
Coers, J. et al. Chlamydia muridarum evades growth restriction by the IFN-γ-inducible host resistance factor Irgb10. J. Immunol. 180, 6237–6245 (2008).
Al-Zeer, M. A., Al-Younes, H. M., Braun, P. R., Zerrahn, J. & Meyer, T. F. IFN-γ-inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PLoS ONE 4, e4588 (2009).
Lapaquette, P., Glasser, A. L., Huett, A., Xavier, R. J. & Darfeuille-Michaud, A. Crohn's disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell. Microbiol. 12, 99–113 (2010).
Nelson, D. E. et al. Chlamydial IFN-γ immune evasion is linked to host infection tropism. Proc. Natl Acad. Sci. USA 101, 10658–10663 (2005).
Bernstein-Hanley, I. et al. The p47 GTPases Igtp and Irgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice. Proc. Natl Acad. Sci. USA 103, 14092–14097 (2006).
Henry, S. C. et al. Impaired macrophage function underscores susceptibility to Salmonella in mice lacking Irgm1 (LRG-47). J. Immunol. 179, 6963–6972 (2007).
Miyairi, I. et al. The p47 GTPases Iigp2 and Irgb10 regulate innate immunity and inflammation to murine Chlamydia psittaci infection. J. Immunol. 179, 1814–1824 (2007).
Singh, S. B. et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nature Cell Biol. 12, 1154–1165 (2010).
Brest, P. et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nature Genet. 43, 242–245 (2011).
Lippmann, J. et al. Dissection of a type I interferon pathway in controlling bacterial intracellular infection in mice. Cell. Microbiol. 13, 1668–1682 (2011).
Hunn, J. P. et al. Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J. 27, 2495–2509 (2008). This paper introduced the concept of regulatory and effector members of the IRG family in the targeting of the pathogen vacuole.
Khaminets, A. et al. Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell. Microbiol. 12, 939–961 (2010).
Cai, Q. & Sheng, Z.-H. Uncovering the role of Snapin in regulating autophagy-lysosomal function. Autophagy 7, 445–447 (2011).
Grégoire, I. P. et al. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog. 7, e1002422 (2011).
McCarroll, S. A. et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nature Genet. 40, 1107–1112 (2008).
Intemann, C. D. et al. Autophagy gene variant IRGM -261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 5, e1000577 (2009).
King, K. Y. et al. Polymorphic allele of human IRGM1 is associated with susceptibility to tuberculosis in African Americans. PLoS ONE 6, e16317 (2011).
Khor, B., Gardet, A. & Xavier, R. J. Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307–317 (2011).
Rupper, A. C. & Cardelli, J. A. Induction of guanylate binding protein 5 by gamma interferon increases susceptibility to Salmonella enterica serovar Typhimurium-induced pyroptosis in RAW 264.7 cells. Infect. Immun. 76, 2304–2315 (2008).
Tietzel, I., El-Haibi, C. & Carabeo, R. A. Human guanylate binding proteins potentiate the anti-Chlamydia effects of interferon-γ. PLoS ONE 4, e6499 (2009).
Modiano, N., Lu, Y. E. & Cresswell, P. Golgi targeting of human guanylate-binding protein-1 requires nucleotide binding, isoprenylation, and an IFN-γ-inducible cofactor. Proc. Natl Acad. Sci. USA 102, 8680–8685 (2005).
Britzen-Laurent, N. et al. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS ONE 5, e14246 (2010).
Nantais, D. E., Schwemmle, M., Stickney, J. T., Vestal, D. J. & Buss, J. E. Prenylation of an interferon-γ-induced GTP-binding protein: the human guanylate binding protein, huGBP1. J. Leukoc. Biol. 60, 423–431 (1996).
Traver, M. K. et al. Immunity-related GTPase M (Irgm) proteins influence the localization of guanylate-binding protein 2 (Gbp2) by modulating macroautophagy. J. Biol. Chem. 286, 30471–30480 (2011).
Virreira Winter, S. et al. Determinants of GBP recruitment to Toxoplasma gondii vacuoles and the parasitic factors that control it. PLoS ONE 6, e24434 (2011). An elegant study demonstrating heterotypic GBP interactions that dictate the targeting of T. gondii by members of this IFN-inducible GTPase family.
Alonso, S., Pethe, K., Russell, D. G. & Purdy, G. E. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc. Natl Acad. Sci. USA 104, 6031–6036 (2007).
Ponpuak, M. et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 32, 1–13 (2010).
Kim, J. Y. & Ozato, K. The sequestosome 1/p62 attenuates cytokine gene expression in activated macrophages by inhibiting IFN regulatory factor 8 and TNF receptor-associated factor 6/NF-κB activity. J. Immunol. 182, 2131–2140 (2009).
Korioth, F., Gieffers, C., Maul, G. G. & Frey, J. Molecular characterization of NDP52, a novel protein of the nuclear domain 10, which is redistributed upon virus infection and interferon treatment. J. Cell Biol. 130, 1–13 (1995).
Lu, F. T. et al. Expression and function of galectin-3, a β-galactoside-binding lectin, in human monocytes and macrophages. Am. J. Pathol. 147, 1016–1028 (1995).
Shahnazari, S. et al. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe 8, 137–146 (2010).
Jabado, N. et al. Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J. Exp. Med. 192, 1237–1248 (2000). This study defined the elusive cation transporter function of NRAMP1 at the phagosomal membrane of macrophages.
Nairz, M. et al. Interferon-γ limits the availability of iron for intramacrophage Salmonella typhimurium. Eur. J. Immunol. 38, 1923–1936 (2008).
White, C., Lee, J., Kambe, T., Fritsche, K. & Petris, M. J. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 284, 33949–33956 (2009).
Wagner, D. et al. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J. Immunol. 174, 1491–1500 (2005).
Govoni, G., Gauthier, S., Billia, F., Iscove, N. N. & Gros, P. Cell-specific and inducible Nramp1 gene expression in mouse macrophages in vitro and in vivo. J. Leukoc. Biol. 62, 277–286 (1997).
Zaharik, M. L. et al. The Salmonella enterica serovar typhimurium divalent cation transport systems MntH and SitABCD are essential for virulence in an Nramp1G169 murine typhoid model. Infect. Immun. 72, 5522–5525 (2004).
Wagner, D. et al. Changes of the phagosomal elemental concentrations by Mycobacterium tuberculosis Mramp. Microbiology 151, 323–332 (2005).
Sibley, L. D. Invasion and intracellular survival of protozoan parasites. Immunol. Rev. 240, 72–91 (2011).
Diefenbach, A. et al. Type 1 interferon (IFNα/β) and type 2 nitric oxide synthase regulate the innate immune response to a protozoan parasite. Immunity 8, 77–87 (1998).
Holscher, C. et al. Defective nitric oxide effector functions lead to extreme susceptibility of Trypanosoma cruzi-infected mice deficient in γ-interferon receptor or inducible nitric oxide synthase. Infect. Immun. 66, 1208–1215 (1998).
Mellouk, S. et al. Nitric oxide-mediated antiplasmodial activity in human and murine hepatocytes induced by gamma interferon and the parasite itself: enhancement by exogenous tetrahydrobiopterin. Infect. Immun. 62, 4043–4046 (1994).
Scharton-Kersten, T. M., Yap, G., Magram, J. & Sher, A. Inducible nitric oxide is essential for host control of persistent but not acute infection with the intracellular pathogen Toxoplasma gondii. J. Exp. Med. 185, 1261–1273 (1997).
Collazo, C. M. et al. Inactivation of LRG-47 and IRG-47 reveals a family of interferon γ-inducible genes with essential, pathogen-specific roles in resistance to infection. J. Exp. Med. 194, 181–188 (2001).
Zhao, Y. et al. Virulent Toxoplasma gondii evade immunity-related GTPase-mediated parasite vacuole disruption within primed macrophages. J. Immunol. 182, 3775–3781 (2009).
Halonen, S. K., Taylor, G. A. & Weiss, L. M. Gamma interferon-induced inhibition of Toxoplasma gondii in astrocytes is mediated by IGTP. Infect. Immun. 69, 5573–5576 (2001).
Martens, S. et al. Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47-resistance GTPases. PLoS Pathog. 1, e24 (2005).
Ling, Y. M. et al. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J. Exp. Med. 203, 2063–2071 (2006).
Zhao, Y., Wilson, D., Matthews, S. & Yap, G. S. Rapid elimination of Toxoplasma gondii by gamma interferon-primed mouse macrophages is independent of CD40 signaling. Infect. Immun. 75, 4799–4803 (2007).
Liesenfeld, O. et al. The IFN-γ-inducible GTPase, Irga6, protects mice against Toxoplasma gondii but not against Plasmodium berghei and some other intracellular pathogens. PLoS ONE 6, e20568 (2011).
Santiago, H. C. et al. Mice deficient in LRG-47 display enhanced susceptibility to Trypanosoma cruzi infection associated with defective hemopoiesis and intracellular control of parasite growth. J. Immunol. 175, 8165–8172 (2005).
Zhao, Z. et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4, 458–469 (2008).
Degrandi, D. et al. Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J. Immunol. 179, 7729–7740 (2007). Together with reference 16, this study provides a comprehensive genomic, molecular and cellular description of the expanded GBP family.
Fentress, S. J. et al. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii secreted kinase promotes macrophage survival and virulence. Cell Host Microbe 8, 484–495 (2010).
Pfefferkorn, E. R. Interferon γ blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc. Natl Acad. Sci. USA 81, 908–912 (1984). This study first showed the importance of tryptophan degradation in restricting the growth of T. gondii.
Carlin, J. M., Borden, E. C. & Byrne, G. I. Interferon-induced indoleamine 2,3-dioxygenase activity inhibits Chlamydia psittaci replication in human macrophages. J. Interferon Res. 9, 329–337 (1989).
Roshick, C., Wood, H., Caldwell, H. D. & McClarty, G. Comparison of gamma interferon-mediated antichlamydial defense mechanisms in human and mouse cells. Infect. Immun. 74, 225–238 (2006).
Daubener, W. et al. Restriction of Toxoplasma gondii growth in human brain microvascular endothelial cells by activation of indoleamine 2,3-dioxygenase. Infect. Immun. 69, 6527–6531 (2001).
Mao, R. et al. Indoleamine 2,3-dioxygenase mediates the antiviral effect of gamma interferon against hepatitis B virus in human hepatocyte-derived cells. J. Virol. 85, 1048–1057 (2011).
Meisel, R. et al. Human but not murine multipotent mesenchymal stromal cells exhibit broad-spectrum antimicrobial effector function mediated by indoleamine 2,3-dioxygenase. Leukemia 25, 648–654 (2011).
Knubel, C. P. et al. Indoleamine 2,3-dioxigenase (IDO) is critical for host resistance against Trypanosoma cruzi. FASEB J. 24, 2689–2701 (2010).
Divanovic, S. et al. Opposing biological functions of tryptophan catabolizing enzymes during intracellular infection. J. Infect. Dis. 205, 152–161 (2012).
Siegrist, F., Eberling, M. & Certa, U. The small interferon-induced transmembrane genes and proteins. J. Interferon Cytokine Res. 31, 183–197 (2011).
Lu, J. et al. The IFITM proteins inhibit HIV-1 infection. J. Virol. 85, 2126–2137 (2011).
Huang, I. C. et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 7, e1001258 (2011).
Weidner, J. M. et al. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 84, 12646–12657 (2010).
Yount, J. S. et al. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nature Chem. Biol. 6, 610–614 (2010).
Feeley, E. M. et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 7, e1002337 (2011).
McNab, F. W., Rasjbaum, R., Stoyle, J. P. & O'Garra, A. Tripartite-motif proteins and innate immune regulation. Curr. Opin. Immunol. 23, 46–56 (2011).
Uchil, P. D., Quinlin, B. D., Chan, W.-T., Luna, J. M. & Mothes, W. TRIM E3 ligases interfere with the early and late stages of the retroviral life cycle. PLoS Pathog. 4, e16 (2008).
Stemlau, M. et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853 (2004). This was the first report of TRIM-mediated activity against HIV-1, which led to the idea that members of the TRIM family can act as intrinsic antiviral factors.
Pertel, T. et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365 (2011). This study adds a second major function for TRIM5 in innate immune signalling cascades following retroviral infection.
Gao, B., Duan, Z., Xu, W. & Xiong, S. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology 50, 424–433 (2009).
Eldin, P. et al. TRIM22 E3 ubiquitin ligase activity is required to mediate antiviral activity against encephalomyocarditis virus. J. Gen. Virol. 90, 536–545 (2009).
Taylor, R. T. et al. TRIM79α, an interferon-stimulated gene product, restricts tick-borne encephalitis virus replication by degrading the viral RNA polymerase. Cell Host Microbe 10, 185–196 (2011).
Mallery, D. L. et al. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21). Proc. Natl Acad. Sci. USA 107, 19985–19990 (2010).
Haller, O. & Kochs, G. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J. Interferon Cytokine Res. 31, 79–87 (2011).
Mordstein, M. et al. Interferon-λ contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 4, e1000151 (2008).
Gao, S. et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465, 502–506 (2010).
Nordmann, A., Wixler, L., Boergeling, Y., Wixler, V. & Ludwig, S. A new splice variant of the human guanylate-binding protein 3 mediates anti-influenza activity through inhibition of viral transcription and replication. FASEB J. 26, 1290–1300 (2012).
Itsui, Y. et al. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 50, 1527–1537 (2009).
Elde, N. C., Child, S. J., Geballe, A. P. & Malik, H. S. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature 457, 485–489 (2009). An insightful study revealing the evolutionary adaptations undergone by PKR to avoid the emergence of viral mimics of its substrate EIF2α.
Harris, R. S. et al. DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809 (2003). The first demonstration of the DNA-deaminating activity of an APOBEC protein against retroviral infection.
Mbisa, J. L., Bu, W. & Pathak, V. K. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J. Virol. 84, 5250–5259 (2010).
Laguette, N. et al. SAMHD1 is the dendritic-and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011).
Hrecka, K. et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011).
Goldstone, D. C. et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 (2011).
Lenschow, D. J. et al. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl Acad. Sci. USA 104, 1371–1376 (2007). This study comprehensively demonstrates the importance of ISG15 as an inhibitory mechanism against phylogenetically diverse viruses.
Durfee, L. A., Lyon, N., Seo, K. & Huibregtse, J. M. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol. Cell 38, 722–732 (2010).
Skaug, B. & Chen Z. J. Emerging role of ISG15 in antiviral immunity. Cell 143, 187–190 (2010).
Bi, Z. & Reiss, C. S. Inhibition of vesicular stomatitis virus infection by nitric oxide. J. Virol. 69, 2208–2213 (1995).
Mannick, J. B., Asano, K., Izumi, K., Kieff, E. & Stamler, J. S. Nitric oxide produced by human B lymphocytes inhibits apoptosis and Epstein-Barr virus replication. Cell 79, 1137–1146 (1994).
Zaragoza, C. et al. The role of inducible nitric oxide synthase in the host response to Coxsackievirus myocarditis. Proc. Natl Acad. Sci. USA 95, 2469–2474 (1998).
Neil, S. J. et al. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–431 (2008). The first report on the role of tetherin in preventing HIV-1 release.
Van Damme, N. et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252 (2008).
Evans, D. T., Serrano-Moreno, R., Singh, R. K. & Guatelli, J. C. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 18, 388–396 (2010).
Perez-Caballero, D. et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139, 499–511 (2009). A well-designed study that demonstrates that the domain structure of tetherin dictates its antiviral tethering activity.
Hinz, A. et al. Structural basis of HIV-1 tethering to membranes by the BST-2/Tetherin ectodomain. Cell Host Microbe 7, 314–323 (2010).
Seo, J. Y., Yaneva, R. & Cresswell, P. Viperin: a multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 10, 534–539 (2011).
Hinson, E. R. & Cresswell, P. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic helix. Proc. Natl Acad. Sci. USA 106, 20452–20457 (2009).
Wang, X., Hinson, E. R. & Cresswell, P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2, 96–105 (2007).
Stzretter, K. J. et al. The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J. Virol. 85, 11557–11566 (2011).
Shapira, S. D. & Hacohen, N. Systems biology approaches to dissect mammalian innate immunity. Curr. Opin. Immunol. 23, 71–77 (2011).
Amit, I., Regev, A. & Hacohen, N. Strategies to discover regulatory circuits of the mammalian immune system. Nature Rev. Immunol. 11, 873–880 (2011).
Aderem, A. et al. A systems biology approach to infectious disease research: innovating the pathogen–host research paradigm. mBio 2, 1–4 (2011).
Bierne, H. & Cossart, P. When bacteria target the nucleus: the emerging family of nucleomodulins. Cell. Microbiol. 23 Feb 2012 (doi:10.1111/j.1462-5822.2012.01758.x).
Schnoor, M., Betanzos, A., Weber, D. A. & Parkos, C. A. Guanylate-binding protein-1 is expressed at tight junctions of intestinal epithelial cells in response to interferon-γ and regulates barrier function through effects on apoptosis. Mucosal Immunol. 2, 33–42 (2009).
Bonazzi, M. & Cossart, P. Impenetrable barriers or entry portals? The role of cell–cell adhesion during infection. J. Cell Biol. 195, 349–358 (2011).
Blanc, M. et al. Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis. PLoS Biol. 9, e1000598 (2011).
Orvedahl, A. et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480, 113–117 (2011).
Pedersen, I. M. et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449, 919–923 (2007).
MacMicking, J. D. Immune control of phagosomal bacteria by p47 GTPases. Curr. Opin. Microbiol. 8, 74–82 (2005).
Gomez, L. M. et al. A polymorphism in the inducible nitric oxide synthase gene is associated with tuberculosis. Tuberculosis 87, 288–294 (2008).
Li, X. L. et al. An essential role for the antiviral endoribonuclease, RNase-L, in antibacterial immunity. Proc. Natl Acad. Sci. USA 105, 20816–20821 (2008).
Acknowledgements
The author apologizes to colleagues whose work has not been cited owing to space constraints. J.D.M. is supported by the US National Institutes of Health (R01AI068041-06), a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award (1007845), a Crohn's and Colitis Foundation of America Senior Research Award (R09928) and a W.W. Winchester Award.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Supplementary information
Supplementary Figure 1
IFN-inducible enzymes in oxidative and nitrosative defense. (PDF 277 kb)
Related links
Glossary
- Autophagy
-
A specialized process involving the degradative delivery of a portion of the cytoplasm or of damaged organelles to the lysosome. Internalized pathogens can also be eliminated by this pathway.
- Reactive oxygen species
-
(ROS). Aerobic organisms derive their energy from the reduction of oxygen. The metabolism of oxygen, and in particular its reduction through the mitochondrial electron-transport chain, generates by-products such as superoxide (O2−) and downstream intermediates such as hydrogen peroxide (H2O2) and hydroxyl radicals (·OH). These three species are referred to as ROS. ROS can damage important intracellular targets, such as DNA, lipids or proteins.
- Reactive nitrogen species
-
(RNS). Nitric oxide (NO) chemistry is complex because of the extreme reactivity of NO, which can result in the formation of different reactive nitrogen intermediates (RNI) depending on the amount of NO that is produced by cells. At low concentrations, NO reacts directly with metals and other radicals. At higher concentrations, indirect effects prevail, and these include several oxidation or nitrosylation reactions with oxygen that result in the production of various congeners. NO and related RNI are effective antimicrobial agents and signal-transducing molecules.
- Phagolysosomes
-
Intracellular vesicles that result from the fusion of phagosomes, which enclose extracellular material that has been ingested, with lysosomes, which contain lytic enzymes and antimicrobial peptides.
- NADPH oxidases
-
Enzyme systems that consist of multiple cytosolic and membrane-bound subunits. The complex is assembled in activated phagocytic cells on the plasma and phagosomal membranes. NADPH oxidase uses electrons from NADPH to reduce molecular oxygen to form superoxide anions. Superoxide anions are enzymatically converted to hydrogen peroxide, which in neutrophils can undergo further conversion by myeloperoxidase to hypochloric acid, a highly toxic and microbicidal agent.
- Respiratory burst
-
The process by which molecular oxygen is reduced by the NADPH oxidase system to produce reactive oxygen species.
- Chronic granulomatous disease
-
An inherited disorder caused by defective oxidase activity in the respiratory burst of phagocytes. It results from mutations in any of five genes that are necessary to generate the superoxide radicals required for normal phagocyte function. Affected patients suffer from increased susceptibility to recurrent infections.
- Galectins
-
Lectins that bind a wide variety of glycoproteins and glycolipids containing β-galactoside. They have extracellular and intracellular functions, including the regulation of apoptosis, RAS signalling, cell adhesion and angiogenesis.
- SNARE proteins
-
(Soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins). A class of proteins that is required for membrane fusion events that occur in the course of vesicle trafficking and secretion.
- ISGylation
-
The attachment of the ubiquitin-like modifier ISG15 to either pathogen or host protein targets to regulate their function rather than stimulate degradation.
- MicroRNAs
-
Single-stranded RNA molecules of approximately 21–23 nucleotides in length that are thought to regulate the expression of other genes.
Rights and permissions
About this article
Cite this article
MacMicking, J. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol 12, 367–382 (2012). https://doi.org/10.1038/nri3210
Published:
Issue date:
DOI: https://doi.org/10.1038/nri3210
This article is cited by
-
TRIM32 reduced the recruitment of innate immune cells and the killing capacity of Listeria monocytogenes by inhibiting secretion of chemokines
Gut Pathogens (2023)
-
Transcriptome analysis of a newly established mouse model of Toxoplasma gondii pneumonia
Parasites & Vectors (2023)
-
Intracellular lifestyle of Chlamydia trachomatis and host–pathogen interactions
Nature Reviews Microbiology (2023)
-
Vγ9+Vδ2+ T cell control of Listeria monocytogenes growth in infected epithelial cells requires butyrophilin 3A genes
Scientific Reports (2023)
-
IFN-γ stimulated murine and human neurons mount anti-parasitic defenses against the intracellular parasite Toxoplasma gondii
Nature Communications (2022)