Abstract
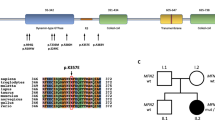
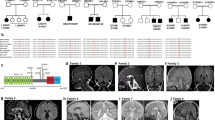
Leukodystrophies are inherited disorders characterized by progressive degeneration of white matter in the central nervous system. Here, we investigate a previously uncharacterized autosomal recessive leukodystrophy which is associated with the homozygous missense variant in ZNF319 (c.800T>C; p.Phe267Ser) in an 18-year-old male presenting with spasticity, ataxia, cognitive decline, and white matter abnormalities on MRI. The variant was absent in population databases (gnomAD, ClinVar) and predicted to be pathogenic by multiple in silico tools. Molecular dynamics simulations revealed that F267 is a stabilizing residue within a β-strand of the zinc finger domain, forming π-stacking and hydrophobic interactions that are lost upon substitution with serine, leading to structural instability, increased flexibility, and protein unfolding. Despite normal transcript and protein expression, ZNF319-F267S mislocalized to the cytoplasm due to disruption of its bipartite nuclear localization signal (NLS), resulting in impaired interaction with importin α1 (KPNA1). Functional analysis confirmed that the mutation disrupts nuclear transport and prevents transcriptional activation of genes involved in myelination. Protein interaction network and gene ontology analysis highlighted ZNF319’s role in transcriptional regulation and its localization in the CHOP-C/EBP transcriptional complex. Expression profiling demonstrated ZNF319 enrichment in oligodendrocytes and white matter regions, correlating with the observed leukoencephalopathy. Our study identifies ZNF319 as a novel gene implicated in human leukodystrophy and highlights how a single-point mutation can compromise nuclear import and transcriptional function, leading to white matter degeneration. These findings expand the genetic landscape of leukodystrophies and provide mechanistic insights into transcriptional regulation in myelin maintenance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data generated or analyzed during this study are available from the corresponding author upon request.
References
van der Knaap MS, Bugiani M. Leukodystrophies: a proposed classification system based on pathological changes and pathogenetic mechanisms. Acta Neuropathol. 2017;134:351–82.
Vanderver A, Prust M, Tonduti D, Mochel F, Hussey HM, Helman G, et al. Case definition and classification of leukodystrophies and leukoencephalopathies. Mol Genet Metab. 2016;119:164–73.
Matthews JM. Zinc fingers: modular protein domains with multiple functions. Biochim Biophys Acta. 2001;1547:116–26.
Mirzaa GM, Chong JX, Piton A, Popp B, Foss K, Guo H, et al. De novo and inherited variants in ZNF292 underlie a neurodevelopmental disorder with features of autism spectrum disorder. Genet Med. 2020;22:538–46.
Stessman HAF, Willemsen MH, Fenckova M, Penn O, Hoischen A, Xiong B, et al. Disruption of POGZ is associated with intellectual disability and autism spectrum disorders. Am J Hum Genet. 2017;98:541–52.
Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–47.
Cassandri M, Smirnov A, Novelli F, Pitolli C, Agostini M, Malewicz M, et al. Zinc-finger proteins in health and disease. Cell Death Discov. 2017;3:17071.
Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Quart Rev Biophys. 2010;43:1–21.
Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74.
Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–40.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9.
DeLano WL. The PyMOL Molecular Graphics System. 2002. https://pymol.org.
Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, et al. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12:255–78.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894.
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539.
Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388.
Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47:D529–D541.
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–9.
Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419.
Papatheodorou I, Moreno P, Manning J, Fuentes AM, George N, Fexova S, et al. Expression Atlas update: from tissues to single cells. Nucleic Acids Res. 2020;48:D77–D83.
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25.
Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods. 2017;14:71–73.
Acknowledgements
We sincerely thank the proband and his family for their participation and continued support for this study.
Funding
The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through Large Research Project under grant number RGP2/257/45.
Author information
Authors and Affiliations
Contributions
S Rehan Ahmad: Conceptualization, study design, experimental execution, data analysis, and manuscript writing. Md. Zeyaullah: Methodology and data analysis. Yousef Zahrani: Genetic counseling and contribution to manuscript preparation. Adam Dawria: Clinical evaluation, diagnosis, and genotype–phenotype interpretation. Abdelrhman AG Altijani: Methodology and data analysis. Ahmed Salih: Methodology and data analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ahmad, S.R., Zeyaullah, M., Zahrani, Y. et al. Pathogenic ZNF319 variant disrupts nuclear localization and transcriptional regulation to cause a novel form of autosomal recessive leukodystrophy. J Hum Genet 70, 577–587 (2025). https://doi.org/10.1038/s10038-025-01386-2
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s10038-025-01386-2