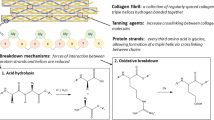
Abstract
A comprehensive evaluation of collagen-based historical artifacts is crucial for the preservation and inheritance of cultural relics, necessitating interdisciplinary approaches that integrate scientific knowledge with practical expertise to develop effective conservation strategies. The present work was focused on a piece of historical leather armor housed in Turpan Museum, Xinjiang, China. SEM–EDS, XRD, ATR-FTIR, and NMR were carried out on this historical leather armor from macro and micro perspectives, mainly including morphology observation, composition analysis, structural assessment, which could provide both quantitative and qualitative insights into the deterioration of the historical leather armor. Additionally, the non-destructive sampling methods and the third-generation sequencing technology (TGS) were employed to identify a total of 13 bacterial species and 8 fungal species, and their metabolic pathways were predicted, providing guidance for preventive conservation and restoration strategies. By the present work, necessary fundamental knowledge might be provided for the effective preservation and inheritance of collagen-based historical artifacts.
Graphical Abstract

Similar content being viewed by others
Introduction
Leather artifacts have carried significant historical and cultural value throughout human history for all nations. However, their deterioration can be influenced by various environment factors, such as humidity, temperature, light, pollution, causing the changes in their appearance, structure and composition [1]. Unlike other cultural relics, such as ceramics and metals, leathers with the main component of collagen are more likely to be affected by environmental factors. Numerous collagen-based artifacts, including the thirteenth-century parchment document [2], shoe fragments discovered in House Page, Germany, and the leather bookbinding from the Qajar era [3], have unfortunately deteriorated by various factors, representing a significant loss. The scarcity and difficulty of preserving leather cultural relics are important issues that cannot be ignored.
In March 2003, a salvage excavation was conducted jointly by the Turpan Cultural Relics Bureau and the Xinjiang Institute of Cultural Relics and Archaeology at the Yanghai Cemetery (geographical coordinates: 42°48′ N, 89°39′ E) [4]. The preservation of leather artifacts in the Turpan Yanghai Cemetery is indeed remarkable due to the unique environmental conditions of the region. Turpan, located in the Xinjiang Uygur Autonomous Region of China, is known for its extremely dry and arid climate, with high temperatures and low humidity levels [5,6,7]. This geographical conditions are conducive to the preservation of organic materials such as leather, as they inhibit the growth of microorganisms and reduce the rate of decomposition. Yanghai Cemetery has yielded a wealth of archaeological finds, as well as various well-preserved leather artifacts, including leather clothing [8], hats, shoes (boots), leather quivers, boxes and bags, leather saddles, bridles, belts, straps, and etc. Most of the tombs exhibited cultural characteristics from the Bronze Age to Iron Age. Among these artifacts, the historical leather armor stands out as a significant discovery. It is noteworthy because it is the only piece of armor excavated from the cemetery, making it a rare and valuable find for researchers and historians. The discovery of the historical leather armor provides valuable insights into ancient military practices, craftsmanship, and the cultural context. The preservation of the historical leather armor in the Turpan Yanghai Cemetery highlights the importance of environmental factors in archaeological preservation, and underscores the significance of a comprehensive evaluation by interdisciplinary approaches. This provides theoretical guidance for preservation efforts.
The common deterioration phenomena of collagen-based cultural relic include decreased hardness, reduced flexibility, loss of structural integrity, accumulation of dust, erosion, and discoloration [9]. The degradation of collagen begins with the breakage of peptide bonds between amino acids, subsequently leading to the releases of varying sizes of peptides from the triple helical structure of collagen [10]. As early as 1988, Michael et. al. conducted an investigation on the potential of amino acid compositions and ratios, such as Asp/Pro ratio, and utilized infrared spectra to determine degraded bone [11]. Up to now, the evaluation of collagen-based artifacts is commonly conducted at macroscopic, microscopic, and molecular levels [9]. Parchment has been extensively investigated, serving as a prominent example in numerous studies on the deterioration of cultural relic. The researches focused on the microscopic observations [12], discoloration [13], thermal behavior [14,15,16], as well as the effects of pollutants [17], microbial attack [18, 19], and temperature-humidity [20]. The similarity in composition between parchment and leather suggests potential similarities in deterioration mechanisms. However, compared to parchment, leather is more complex in terms of process preparation and composition. Since this historical leather armor has not undergone a systematic assessment since its excavation yet, progress in preservation and restoration work has been limited. Therefore, there are still many gaps that need to be filled in the scientific deterioration evaluation.
This study conducted a comprehensive evaluation of a historical leather armor through morphology observation, composition analysis, structural assessment, and microbial identification and prediction, providing valuable insights into the historical and cultural significance of the collagen-based artifacts.
Material and methods
The historical leather armor
The tomb with the unearthed leather armor (number: IIM127:11–2) was in the western part of Cemetery II. A plan of Yanghai II cemetery with the precise excavation location of the historical leather armor is presented in Fig. S1 [4]. The tomb measured 1.65 m in length, 0.84 m in width, and 1.32 m in depth. The unearthed leather armor is relatively rare worldwide, and this piece is the only armor found among the thousands of tombs excavated in the Yanghai Cemetery. The full-view digital image and detailed images are shown in Fig. 1a–e. The entire leather armor was composed of over 5,000 leather pieces, crafted from two different shapes and sizes of high-quality cowhide. The short strip leather is a curved top rectangle measuring \(2.5 * 1.5\) cm, while the long strip leather is a rectangle measuring \(7.8 * 1.6\) cm. The connection between these strip leathers is shown in Fig. S2 [4].
SEM analysis
The Quanta 250 scanning electron microscope (FEI Company) was utilized to acquire micrographs of the grain layer and cross-section of the historical leather armor. The imaging process was conducted at an accelerating voltage of 20 kV. Before observation via SEM, the samples were sprayed with a thin layer of gold to enhance their conductivity and imaging quality. EDS was conducted using Quantax 200 XFlash 6/60 (Bruker).
XRD analysis
The XRD analysis was conducted using an X-ray diffractometer (Rigaku Ultima IV, Rigaku Corporation, Tokyo, Japan) with a scanning range from 5° to 90° at a scanning rate of 10°/min. Three XRD analyses were conducted on the soil surrounding the historical leather armor excavated from the site. And the three samples were obtained from three different sampling locations, with two taken from around the burial site and one from the surface of the armor.
ATR-FTIR analysis
The Nicolet iS5 spectrometer (Thermo Fisher Scientific, Madison, WI, USA) with ATR accessories was employed to analyze the historical leather armor and new leather. Spectra were collected in the wavenumber range of 400–4000 cm−1, with 32 scans and a spectral resolution of 4 cm−1.
Solid state NMR
The solid-state 13C CP/MAS NMR measurement cross-polarization/magic angle spinning (CP/MAS) experiment was conducted by an Agilent 600 DD2 spectrometer (Agilent, USA) with the magnetic field strength of 14.1 T at Larmor frequency of 150.72 MHz for 13C. The experiment employed a 4 mm double-resonance 1H-X MAS probe operating at a MAS frequency of 8 kHz, with proton decoupling (TPPM) applied during acquisition. And a relaxation delay of 3 s was utilized, along with a CP contact time of 2 ms and an approximately 9090 during acquisitionμs). The data were acquired through 914 scans. The reference for the 13C chemical shift was established using the signals from Adamantane at 38.4 and 29.5 ppm.
Microbial identification
The third-generation sequencing technology (TGS) can achieve megabase-level long-read sequencing and non-amplification-based genome modification detection by DNA extraction, PCR and 16S rRNA/ITS gene amplicon sequencing. The sampling point for microbial analysis was circled in yellow in Fig. 1b. The rationality of the method and the accuracy of the data volume were confirmed in Table S1 and Fig. S3.
Metabolic pathways analysis
Microbial pathways analysis involves aligning known microbial genomic sequences to construct an evolutionary tree and infer ancestral gene functional profiles. Castor hidden state prediction algorithm was used to determine gene family copy numbers for feature sequences. Abundance data of metabolic pathways in each sample were obtained through Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) analysis and metabolic pathway analysis.
Results and discussion
Morphology observation
The scanning electron microscope (SEM) was used to observe the microstructure of the detached fragments of the grain layer (Fig. 2a) and cross-section (Fig. 2b) of the historical leather armor. In the SEM images, the collagen fibers in the historical leather armor are relatively long and thick, with diameters ranging from 3 to 30 μm. The fibers are twisted into bundles, and surface samples present typical structural characteristics of cowhide leather when compared with new leather’s microscopic features (Fig. S4). The fiber bundles’ surface has rough granules, and at higher magnifications, the scanning electron microscope images reveal a dense coating. And the typical periodic D-band structure of collagen cannot be observed.
Figure 3 iluustrates the SEM–EDS images of the grain layer and cross-section of the historical leather armor. Through EDS mapping results, it can be found that the distribution of the elements C, N, and O in the fibers is consistent with the fiber morphology. While the elements of Fe, Mn, Si, and Al are uniformly distributed on the sample’s surface, which might be attributed to its burial environment. Given that, the source of these unidentified contaminants were further confirmed by the following component analysis based on the XRD results.
Component analysis
The XRD analysis was used for the specific identification of the contaminants from soil. The results are shown in Fig. 4. The predominant minerals in the two soil samples taken surrounding the burial were quartz, albite, and coesite. The other soil sample collected from the historical leather armor indicated the presence of quartz, albite, magnetite, hedenbergite, and siderite. These minerals are widely distributed natural components found on the Earth’s surface, and fit well with the elements detected by the results of EDS and XRF (as illustrated in Table S2), thereby confirming our hypothesis.
Given that the elements of C, N, and O are the primary elements in EDS, we conducted an analysis of the organic components in the historical leather armor. The normalized ATR-FTIR spectra of the new leather and the historical leather armor were obtained, as shown in Fig. 5a. A broad peak was observed in the range of 3650 to 2950 cm−1, which was ascribed to the stretching vibrations of -OH and N–H groups. The amide I peaks of the new leather and the historical leather armor appeared at 1641 and 1645 cm−1, respectively, should be associated with the carbonyl stretching, with contributions from the C-N stretching and N–H bending. The amide II peaks of the new leather and the historical leather armor were observed at 1556 and 1549 cm−1, respectively, corresponding to the combination of bending vibrations of the peptide bond (C-N stretch) and in-plane bending vibrations of N–H. The Amide III band peaks of the new leather and the historical leather armor appeared at 1240 and 1242 cm−1, respectively, which should be related to the coupling between the C-N stretch and N–H bend, with contributions from the C=O in-plane bend and C–C stretch [21,22,23,24]. Additionally, the amino acid composition of collagen is illustrated in Fig. 5b. Glycine (Gly), proline (Pro), and hydroxyproline (Hyp), as the characteristic amino acids of collagen protein, collectively constitute over 40% of the total amino acid content. Therefore, the main component of this historical leather was identified as collagen based on the distinct characteristic peaks observed in FTIR and the presence of characteristic amino acid components.
Structure assessment
Information regarding the structure of collagen can also be obtained at the molecular level from the FTIR spectrum (Fig. 5a). Taking an example of the amide I band, the conformation-dependent changes of it are in close relative with intensity. This region corresponds to the vibration of amide carbonyls along the polypeptide backbone [25]. The complexity of the amide I band arises from the interplay between carbonyl stretching modes and the heterogeneity among the backbone carbonyl groups [26]. The new leather sample exhibits two distinct peaks at 1665 cm−1 and 1640 cm−1, respectively, whereas there are no prominent peaks between 1660 and 1670 cm−1 for the historical leather armor sample. The band at 1660 cm−1 is associated with the collagen native triple helix [25], indicating the damage in the internal triple helical structure of the historical leather armor. In addition, the ratio of the peak intensity of the amide III band to the peak at around 1450 cm−1 (PeakAmid III /Peak1450 cm−1) could reflect the integrity of the collagen triple helix structure [27,28,29,30]. The closer the ratio to 1, the more intact the triple helix structure of the collagen. Here, the ratio for new leather is 0.87, which is only 0.35 for the historical leather armor, confirming that the appearance of the deterioration in the historical leather armor.
The deterioration was confirmed using NMR analysis, according to Zhang’s report [31] related closely to the historical leather artifacts. Figure 6 presents the 13C CP/MAS spectrum of the historical leather armor. The 13C signal within 180–160 ppm are assigned to the carbohyl carbon. The signals corresponding to the aromatic carbons in Region I are not observed [32]. The 13C resonant lines in the range of 90–0 ppm in region II contain the peptide aliphatic carbon, which contributes to the amino acid carbon resonances. The peaks at 73.3, 44.6, and 30.4 ppm are attributed to C-4 of hydroxyproline (Hyp), C-2 of glycine (Gly), and C-3 of proline (Pro), respectively [33]. The intensities of Hyp, Gly, and Pro are used as indicators to identify the effects of collagen degradation because collagen has a triple helix structure dominated by the amino acid sequence of Gly-Pro-X or Gly-X-Hyp triplets [34]. Taking the carbonyl carbon peak at 175.9 ppm as a reference peak, the relative fraction of Hyp, Gly, and Pro were 26.3%, 39.1%, and 29.5%, respectively. The results by NMR are consistent to the results of the amino acid content in Fig. 6b. For the newly prepared leather, the relative fraction of Hyp, Gly, and Pro were reported approximately 38%, 35%, and 45% [31]. So the Hyp and Pro in the historical leather armor degraded significantly. The degree of degradation level of the historical leather armor is close to that reported for artifact No. N96 [31], also from Xinjiang, but from different cemeteries.
Additionally, vegetable tanning and alum tanning are recognized as the most ancient methods of leather treatment. Considering the absence of a distribution pattern along the fiber morphology for Al (Fig. 4), and the lack of characteristic aromatic peaks of tannins in Fig. 6, it is plausible to suggest that the historical leather armor might be either untanned or that the tannins present in the leather have degraded due to deterioration or an absence of tannin treatment on this ancient leather armor.
Microbial identification and metabolic pathway analysis
Through gene identification and matching, a total of 13 bacterial and 3 fungal species were detected on the surface of the leather armor artifacts, as listed in Table 1. Among the bacterial species, one was identified at the phylum, class, and order levels, two at the family level (Bacillaceae and Planococcaceae), and six at the genus level (Bacillus, Oceanobacillus, Priestia, Pseudogracilibacillus, Siminovitchia, and Virgibacillus). Among them, Bacillus and Priestia were the dominant bacteria, constituting the main bacterial components of the microbial community on the artifacts’ surface. At the species level, 13 bacterial species were identified, including Bacillus sp. (in: firmicutes), Bacillus sp. 1002B12 12ECASO, Bacillus sp. 81C102Y12, Oceanobacillus caeni, Priestia endophytica, Priestia megaterium, Pseudogracilibacillus sp., Siminobitchia farraginis, Siminovvitchia fordii, Virgibacillus oceani, Virgibacillus sp. WSR35, Paenisporosarcina sp. BE572, and Sporosarcina koreensis. Among them, Bacillus sp. (in: firmicutes) and Priestia endophytica were identified as the dominant species. Regarding fungal composition, Ascomycota was identified at the phylum level, and Eurotiomycetes, Saccharomycetes, and Dothideomycetes were identified at the class level. At the order level, Eurotiales, Saccharomycetales, and Capnodiales were detected. At the family level, Aspergillaceae and Pichiaceae were identified. At the genus level, Aspergillus and Pichia were detected. Finally, at the species level, 3 fungal species were identified: Aspergillus piperis, Aspergillus subversicolor, and Pichia garciniae. Compared to the microbial species extracted from other artifacts [35,36,37,38,39], the historical leather armor unearthed from Turpan contains noticeably fewer microbial species, possibly due to the unique geographical environment of the region. Turpan has a mean annual precipitation of only 16.4 mm but experiences evaporation of 3000 mm, and the temperatures range from around − 9 °C in the coldest months and approximately 45.6 °C in the hottest months [5,6,7], which may not be conducive to microbial growth and metabolic activity.
Figure 7 shows the abundance of MetaCyc secondary functional pathways for bacteria and fungi. These metabolic pathways are classified into 6 major categories, and each category of metabolic pathways is further divided into multiple levels. The biosynthesis of bacteria is mainly the biosynthesis of amino acid, while fungal biosynthesis is predominantly focused on fatty acid and lipid biosynthesis. In comparison to bacteria, fungi exhibit a superior capacity for carbohydrate synthesis, but demonstrate relatively weaker abilities in amino acid biosynthesis. The function of degradation, utilization, and assimilation are crucial in the context of organic carbon accumulation [40]. Among these functions, both bacteria and fungi possess amino acid degradation as well as C1 compound utilization and assimilation capabilities. However, fungi lack the ability to degrade aromatic compound, carboxylate, and secondary metabolite. In term of the generation of precursor metabolites and energy, fungi exhibit the capacity for chitin degradation to ethanol and methyl ketone biosynthesis, which is not observed in bacteria. Conversely, bacteria can perform formaldehyde oxidation and photosynthesis, which are not observed in fungi. Furthermore, compared to bacteria, fungi demonstrate significantly enhanced proficiency in glycan biosynthesis. The preservation environment for artifacts after excavation differs significantly from their burial environment. Therefore, understanding the types of microbes present on the surface of these artifacts and their metabolic pathways is crucial for their long-term preservation.
Conclusion
In this work, a historical leather armor housed in Turpan Museum was evaluated through four aspects: morphology observation, composition analysis, structural assessment, and microbial identification and prediction. The uses of FTIR and NMR as effective tools in the assessment of deterioration has been demonstrated. And the main component and contamination were identified respectively, providing scientific foundation for the reasonable preservation and restoration of collagen-based historical artifacts.
Availability of data and materials
The ancient leather armor is currently housed in the Turpan Museum, located in Xinjiang, China. The data used in the present study are available from the corresponding author upon reasonable request.
References
Abdel-Maksoud G, Abdel-Nasser M, Sultan MH, Eid AM, Alotaibi SH, Hassan SE, Fouda A. Fungal biodeterioration of a historical manuscript dating back to the 14th century: an insight into various fungal strains and their enzymatic activities. Life (Basel). 2022. https://doi.org/10.3390/life12111821.
Lech T. Evaluation of a parchment document, the 13th century incorporation charter for the city of krakow, poland, for microbial hazards. Appl Environ Microbiol. 2016;82:2620–31.
Ershad-Langroudi A, Mirmontahai A. Thermal analysis on historical leather bookbinding treated with PEG and hydroxyapatite nanoparticles. J Therm Anal Calorim. 2015;120:1119–27.
C. Sun. Turfanological Research. Xinjiang Tulufan Society. 2005;(2).
Du L, Wong JS, Li Z, Chen L, Zhang B, Lei B, Peng Z. Hydroclimatic Change in Turpan Basin under Climate Change. Water. 2023;15:3422.
Petraglia MD, Tang Y-N, Li X, Yao Y-F, Ferguson DK, Li C-S. Environmental reconstruction of Tuyoq in the fifth century and its Bearing on Buddhism in Turpan, Xinjiang China. PLoS ONE. 2014;9: e86363.
Yang L, Fu R, He W, He Q, Liu Y. Adaptive thermal comfort and climate responsive building design strategies in dry-hot and dry-cold areas: Case study in Turpan China. Energy Build. 2020;209:109678.
Zhang M, Zhang Z, Wang F, Liu J, Lei Y, Albu Kaya MG, Tang K. A novel identification method for collagen-based cultural heritage: Integrating thermokinetics and generalized master plots.J. Cult Herit. 2024;67:226–36.
R.L. Kathleen Mühlen Axelsson, Dorte V.P. Sommer, Rikke Melin. Degradation of collagen in parchment under the influence of heat-induced oxidation: Preliminary study of changes at macroscopic, microscopic, and molecular levels. Stud Conserv. 2014. https://doi.org/10.1179/2047058414Y.0000000140.
Matthew MSR, Collins J, Child AM, Walker GT. A basic mathematical simulation of the chemical degradation of ancient collagen. J Archaeol Sci. 1995;22:175–83.
Michael SW, Deniro J. Chemical, enzymatic and spectroscopic characterization of “collagen” and other organic fractions from prehistoric bones. Geochim Cosmochim Ac. 1988;52:2197–206.
Bicchieri M, Biocca P, Colaizzi P, Pinzari F. Microscopic observations of paper and parchment: the archaeology of small objects. Herit Sci. 2019;7:1–12.
Pinar G, Sterflinger K, Pinzari F. Unmasking the measles-like parchment discoloration: molecular and microanalytical approach. Environ Microbiol. 2015;17:427–43.
Sebestyén Z, Czégény Z, Badea E, Carsote C, Şendrea C, Barta-Rajnai E, Bozi J, Miu L, Jakab E. Thermal characterization of new, artificially aged and historical leather and parchment. J Anal Appl Pyrol. 2015;115:419–27.
Axelsson KM, Larsen R, Sommer DVP, Melin R. Degradation of collagen in parchment under the influence of heat-induced oxidation: preliminary study of changes at macroscopic, microscopic, and molecular levels. Stud Conserv. 2016;61:46–57.
Budrugeac P, Miu L. The suitability of DSC method for damage assessment and certification of historical leathers and parchments. J Cult Herit. 2008;9:146–53.
Ciglanská M, Jančovičová V, Havlínová B, Machatová Z, Brezová V. The influence of pollutants on accelerated ageing of parchment with iron gall inks. J Cult Herit. 2014;15:373–81.
Cicero C, Pinzari F, Mercuri F. 18th Century knowledge on microbial attacks on parchment: Analytical and historical evidence. Int Biodeter Biodegr. 2018;134:76–82.
Kraková L, Chovanová K, Selim SA, Šimonovičová A, Puškarová A, Maková A, Pangallo D. A multiphasic approach for investigation of the microbial diversity and its biodegradative abilities in historical paper and parchment documents. Int Biodeter Biodegr. 2012;70:117–25.
Badea E, Della Gatta G, Usacheva T. Effects of temperature and relative humidity on fibrillar collagen in parchment: a micro differential scanning calorimetry (micro DSC) study. Polym Degrad Stabil. 2012;97:346–53.
G.J.v. Klinken. Bone collagen quality indicators for palaeodietary and radiocarbon measuurements. J Archaeol Sci. 1999;26:687–95.
Boyatzis SC, Velivasaki G, Malea E. A study of the deterioration of aged parchment marked with laboratory iron gall inks using FTIR-ATR spectroscopy and micro hot table. Herit Sci. 2016. https://doi.org/10.1186/s40494-016-0083-4.
Barth A. Infrared spectroscopy of proteins. Biochim Biophys Ac. 2007;1767:1073–101.
E.R.B. Barbara Brodsky Doyle. Infrared spectroscopy of collagen and collagen-like polypeptides. Biopolymers. 1975;14:937–57.
Rabotyagova OS, Cebe P, Kaplan DL. Collagen structural hierarchy and susceptibility to degradation by ultraviolet radiation. Mater Sci Eng C Mater Biol Appl. 2008;28:1420–9.
Sionkowska A. Effects of solar radiation on collagen and chitosan films. J Photochem Photobiol B. 2006;82:9–15.
He L, Mu C, Shi J, Zhang Q, Shi B, Lin W. Modification of collagen with a natural cross-linker, procyanidin. Int J Biol Macromol. 2011;48:354–9.
Alireza Koochakzaei HA, Mallakpour S. An experimental comparative study of the effect of skin type on the stability of vegetable leather under acidic condition. J Am Leather Chem As. 2018;113:345–51.
Sizeland KH, Hofman KA, Hallett IC, Martin DE, Potgieter J, Kirby NM, Hawley A, Mudie ST, Ryan TM, Haverkamp RG, Cumming MH. Nanostructure of electrospun collagen: Do electrospun collagen fibers form native structures? Materialia. 2018;3:90–6.
Cosma DV, Tudoran C, Coros M, Socaci C, Urda A, Turza A, Rosu MC, Barbu-Tudoran L, Stanculescu I. Modification of cotton and leather surfaces using cold atmospheric pressure plasma and TiO2-SiO2-reduced graphene oxide nanopowders. Materials (Basel). 2023. https://doi.org/10.3390/ma16041397.
Zhang Y, Liu X, Wang Y, Tang H, Qu L, Shang Y, Chen W. Quantitative assessment of collagen degradation in archeological leather by solid-state NMR. J Cult Herit. 2022;58:179–85.
Proietti N, Di Tullio V, Carsote C, Badea E. 13C solid-state NMR complemented by ATR-FTIR and micro-DSC to study modern collagen-based material and historical leather. Magn Reson Chem. 2020;58:840–59.
Aliev AE. Solid-state NMR studies of collagen-based parchments and gelatin. Biopolymers. 2005;77:230–45.
R Larsen, J Wouters, C Chahine, C Calnan, P Brimblecombe. Recommendations on the production, artificial ageing, assessment, storage and conservation of vegetable tanned leather, In Environment Leather Project: Deterioration and Conservation of Vegetable Tanned Leather. 1996;189–202.
Guadalupe P, Sterflinger K, Ettenauer J, Quandt A, Pinzari F. A combined approach to assess the microbial contamination of the Archimedes Palimpsest. Microb Ecol. 2015;69:118–34.
Domenico P, Buterflinger M, Ettenauer J, Quandt A, Pinzari F. A combined approach to assess the microbial contamination of the Archimedes Palimpsest. Microb Ecol. 2015;69:118–34.
Liu Z, Wang Y, Pan X, Ge Q, Ma Q, Li Q, Fu T, Hu C, Zhu X, Pan J. Identification of fungal communities associated with the biodeterioration of waterlogged archeological wood in a Han dynasty tomb in China. Front Microbiol. 2017;8:1633.
Beata G, Celikkol-Aydin S, Bonifay V, Otlewska A, Aydin E, Oldham AL, Brauer JI, Duncan KE, Adamiak J, Sunner JA, Beech IB. Metabolomic and high-throughput sequencing analysis-modern approach for the assessment of biodeterioration of materials from historic buildings. Front Microbiol. 2015;6: 154203.
Liu Z, Zhang Y, Zhang F, Hu C, Liu G, Pan J. Microbial community analyses of the deteriorated storeroom objects in the Tianjin Museum using culture-independent and culture-dependent approaches. Front Microbiol. 2018;9:802.
Qi JY, Yao XB, Lu J, He LX, Cao JL, Kan ZR, Wang X, Pan SG, Tang XR. A 40 % paddy surface soil organic carbon increase after 5-year no-tillage is linked with shifts in soil bacterial composition and functions. Sci Total Environ. 2023;859: 160206.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (No. 52073262, 52373109), National Key R&D Program of China (No. 2017YFB0308500), and Science and Technology Department of Henan Province, China (No. 232102521017).
Author information
Authors and Affiliations
Contributions
MZ was primarily responsible for conducting the investigation and analysis, which included experimental design, data analysis, diagram creation, and manuscript writing. JF carried out most of the SEM and SEM–EDX analyses. JL took responsible for visualization and validation. YC, YL, and YL were in charge of sampling and preservation of leather artifacts. MA was assigned with the task of reviewing and revising the manuscript. KT played a pivotal role in supervision, resources, and funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, M., Fan, J., Liu, J. et al. A comprehensive evaluation of a historical leather armor from Yanghai Cemetery, Turpan. Herit Sci 12, 162 (2024). https://doi.org/10.1186/s40494-024-01275-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1186/s40494-024-01275-5
Keywords
This article is cited by
-
Revealing the secrets of a lacquered leather artifact through molecular fingerprints
npj Heritage Science (2025)
-
Quantitative assessment of hardened leather artifact deterioration using infrared spectroscopy
Heritage Science (2024)