Abstract
The molecular etiology of numerous risk genes for autism spectrum disorder (ASD), including Cadherin 11 (CDH11), remains elusive. We investigated the role of CDH11 in the development of ASD-related behaviors using gene-deficient mice. CDH11 is enriched at synapses in glutamatergic neurons of the anterior cingulate cortex (ACC), which project to the dorsal striatum, nucleus accumbens, and the basolateral amygdala. Developmental ablation of Cdh11 in these neurons led to increased self-grooming, reduced sociability, and decreased neuronal activity in the ACC. Chemogenetic inhibition of ACC glutamatergic neurons recapitulates the over-grooming phenotype, while activation of these neurons mitigates self-grooming in Cdh11-deficient mice. Moreover, targeted expression of CDH11 in the ACC of these mice significantly attenuated their over-grooming behaviors. Proteomics of ACC synaptosomes and CDH11 interactomes revealed that CDH11 participates in synaptic vesicle trafficking, as evidenced by a reduction in presynaptic vesicle density at excitatory synapses in Cdh11-deficient mice. These findings highlight an important role of CDH11 in the development of ASD-related brain circuits and provide insights into the molecular basis of repetitive behaviors in ASD.

Similar content being viewed by others
Introduction
Autism spectrum disorder (ASD) is a complex neurodevelopmental condition characterized by impaired social interaction, compromised verbal and non-verbal communication, and restricted interests or repetitive behaviors [1]. The etiology of ASD is multifaceted, involving genetic, environmental, and neurobiological factors. The genetic landscape of ASD is remarkably diverse, with numerous risk genes contributing to its complex pathology to varying extents [2,3,4]. High-throughput genomic studies have led to the discovery of a wide array of genetic variations, from single nucleotide polymorphisms to copy number variations, that are implicated in ASD [5, 6]. These genes are involved in various developmental processes, including neurogenesis, neuronal migration, axon and dendrite development, and synaptogenesis [7,8,9,10]. Notably, many of these risk genes are prominently expressed in brain regions that are pivotal for behaviors and functions commonly impaired in ASD, such as the prefrontal cortex, amygdala, striatum, hippocampus, and cerebellum [1, 11]. This expression pattern underscores a potential mechanistic link between genetic disruptions and the behavioral phenotypes observed in ASD. The convergent expression of these genes in brain regions associated with ASD suggests a shared pathway or circuit vulnerable to disruption, leading to the characteristic features of ASD [12,13,14].
A group of high-confidence risk genes for ASD encodes synaptic adhesion molecules, such as cadherins, neuroligins, neurexins, and contactin-associated protein [15, 16]. These genes emphasize the crucial role of synaptic adhesion molecules in the development and function of ASD-related neural circuits. Disruptions in synaptic adhesion can lead to altered synaptic connectivity, likely contributing to the behavioral and cognitive symptoms observed in individuals with ASD. Among the numerous adhesion molecules, Cadherin 11 (CDH11) is recognized as an ASD-associated gene [17,18,19,20]. CDH11 encodes a type II classical cadherin from the cadherin superfamily, integral to homophilic cell-cell adhesion. Genetic studies have highlighted the significant roles of CDH11 gene mutations in psychiatric and affective disorders, extending beyond ASD to other conditions such as, bipolar disorder, schizophrenia, and major depressive disorder [21, 22], underscoring its broader impact on brain development and function. However, prior research on CDH11 has predominantly explored its roles in cancer and the immune system, focusing on its contributions to cell adhesion and migration, and its implications for tumor progression and inflammatory responses [23].
Several lines of experimental studies indicate the important roles of CDH11 in brain development. Aberrant expression of CDH11 has been observed in neuronal cultures derived from autistic patients [18]. Cdh11 and another cadherin family member, Cdh9, are expressed at high levels in complementary patterns in sub-populations of Purkinje cells within ASD-relevant regions of the cerebellum, suggesting their functional involvement in ASD [11]. The knockdown of Cdh11 has been shown to impair synapse development in neuronal cultures [24]. Cdh11 gene knockout mice exhibit autism-like behavioral deficits, including reduced sociability, increased self-grooming, and impaired motor coordination [17, 25]. However, the molecular and circuit mechanisms through which CDH11 mutations lead to ASD-related behavioral deficits remain largely unknown.
In the present study, we employed multidisciplinary approaches to investigate how CDH11 affects the development and function of ASD-related brain circuits using Cdh11-deficient mice. Our findings reveal that CDH11 is enriched at synapse of ASD-associated brain regions including ACC and is essential for the trafficking of presynaptic vesicles, with deficits in this function leading to repetitive behaviors. This study provides new insights into the molecular mechanisms underlying the repetitive behaviors associated with ASD.
Materials and methods
Animals
The Cdh11-flox mouse allele was generated by Cyagen Biosciences (TKC-160218-AJH-01, Suzhou, Jiangsu, China) on a C57BL/6J genetic background. The Emx1-Cre and Dlx5/6-Cre mice were obtained from the Jackson Laboratory (Maine, USA), the CamKIIα-Cre mice were provided by Dr. Dongmin Yin (East China Normal University), and the Vasa-Cre mice were provided by Dr. Yuqiang Ding (Fudan University). Cdh11-flox mice were crossbred with Vasa-Cre, Emx1-Cre, CamKIIα-Cre, and Dlx5/6-Cre mice, respectively, to produce general gene knockout and lineage-specific conditional knockout mice. Wild-type C57BL/6J was purchased from Shanghai Jihui Experimental Animal Breeding Co., Ltd (Shanghai, China). All animals used in the experiment were housed in the Specific Pathogen-Free level rodent center at the Institute of Brain Functional Genomics of East China Normal University. Animals were housed 3-5 per cage at 22 ± 2 °C with a 12-hour light/dark cycle, lights on at 7:00 AM and off at 7:00 PM, with food and water provided ad libitum. All animal procedures were approved by and conducted under the supervision of the Institutional Animal Care and Use Committee of East China Normal University (m20221105).
Behavior tests
Adult male mice aged 2–3 months were used for behavioral tests. The experimenter handled the mice for 5 min per day for three consecutive days before the behavior tests. For all behavioral tests, both the experimentation and data analysis were conducted by an experimenter blinded to the mouse genotype and treatment whenever possible. Only male mice were analyzed in behavioral tests, as preliminary studies indicated that female mice exhibited milder behavioral phenotypes.
Grooming analysis
Self-grooming behavior was observed using the HomeCageScan system (CleverSys, Inc., VA, USA). The test mouse was placed in a cage resembling its natural living environment to accurately reflect spontaneous activities. The experimental area was soundproofed from the outside, with uniform illumination at a brightness of 13 lux, clean bedding, and sufficient water and food. After a 10-min acclimation period, the activity of the mice was recorded using a camera for 15 min. Self-grooming bouts and the total time spent in self-grooming of mice were measured based on the manual review of the video recordings.
To observe water spraying-induced grooming behavior, the subject mouse was placed in a cage to adapt for 10 min. The experimenter then sprayed water (23 °C) onto the back of the mouse eight times until the back was fully wetted. Mouse activities were video recorded for 10 min. The grooming bouts and the total grooming time were measured based on the video review.
Three-stage sociability test
Before the sociability test, the subject mouse was placed in a cage for habituation for a 15 min habituation period. Stage 1: the subject mouse was allowed to interact with a novel male mouse (Stranger 1) for 3 min in the same cage. Rest period: the subject mouse was placed back in its home cage to rest for 30 min. Stage 2: the subject and Stranger 1 were placed in the same cage again for 3 min. Rest period: the subject was placed back in its home cage to rest for another 30 min. Stage 3: the subject was allowed to interact with another novel male mouse (Stranger 2) for 3 min in the same cage. The activities of the mice were video recorded, and the frequency and total duration of social interaction of the subject in each stage were manually measured based on the video recordings.
Fear conditioning test
The experiment was conducted using the Startle and Fear Conditioning System (Panlab, Barcelona, Spain). From day 1 to day 3, the mouse was placed in Box A for 5 min each day for adaptation. On day 4, 3 min after entering Box A, the mouse received a tone stimulation (30 s, 2000 Hz, 85 dB), and a foot shock (0.75 mA, 2 s) was delivered during the last 2 seconds of the tone (co-termination). On day 5, the contextual fear test was performed in Box A without any stimulation. The cued fear test was performed in Box B, which had a different wall color from Box A. The subject mouse received a tone stimulation (30 s, 2000 Hz, 85 dB) 3 min after entering Box B. The fear level of the mouse was measured by the freezing time, which was recorded by the system.
Open field test
The subject mouse was placed in a square arena (26 × 26 × 37 cm, W × D × H) for free movement for 30 min. A central 16 × 16 cm region was designated as the central zone, while the remaining space was defined as the peripheral zone. The total locomotion distance and the time spent in both the central and peripheral zones were analyzed using the Tru-Scan photo beam tracking system (Coulbourn Instruments, PA, USA).
Immunohistochemistry
Mice were deeply anesthetized with 2% sodium pentobarbital in ddH2O (20 mg/kg) 1.5 h after the self-grooming test and transcardially perfused with 4% paraformaldehyde solution in PBS. The brains were subsequently dehydrated in 20% and 30% sucrose in PBS for 36 h each, and then sectioned coronally at a thickness of 40 μm using a −20 °C cryostat (Leica, CM3050S, Germany). Five to six sections per mouse, covering the ACC region (A/P +1.1 mm to +0.3 mm from bregma), were used for c-Fos immunostaining. The sections were incubated at 4 °C for 72 h with a primary antibody against c-Fos (rabbit, 1:750, CST #2250S; diluted in blocking solution containing 5% goat serum and 0.3% Triton X-100). Subsequently, the sections were incubated at room temperature for 2 h with a secondary antibody (goat-anti-rabbit IgG conjugated with Alexa Fluor 488, 1:1000, #A-11029, Thermo Fisher). Between antibody incubation steps, samples were rinsed in PBS 3 to 4 times. Tile-scan images covering the ACC region were acquired using a fluorescence microscope at 10x magnification (Leica, DM6B). The number of c-Fos+ nuclei was counted by the observers blinded to the treatment.
In situ hybridization
Chromogenic in situ hybridization was performed using the RNAscope® 2.5 HD Detection Reagent-Brown kit (Advanced Cell Diagnostics #322310-USM, USA) according to the manufacturer’s instructions. Fresh mouse brain tissues (E17, P5, and P14) were fixed in 10% neutral formalin for 24–36 h at room temperature (RT). The samples were then dehydrated through a graded ethanol series, cleared in xylene, and embedded in paraffin. Embedded tissues were sectioned at 5-μm using a microtome (RM2245, Leica) and baked at 60 °C for 1 h. The sections were deparaffinized and preprocessed, including treatment with hydrogen peroxide for 10 min at RT, target retrieval for 15 min at 99 °C, and protease treatment for 30 min at 40 °C. The Cdh11 probe (#506661, ACD) was hybridized to the brain sections for 2 h at 40 °C. Post-hybridization, signal amplification was carried out sequentially with the application of multi-channel fluorescence Amp 1 (30 min at 40 °C), Amp 2 (15 min at 40 °C), Amp 3 (30 min at 40 °C), Amp 4 (15 min at 40 °C), Amp 5 (30 min at RT), Amp 6 (15 min at RT). Diaminobenzidine (DAB-A:DAB-B = 1:1) was added to the slices and incubated at RT for 10 min. The slides were then stained with 50% Hematoxylin for 2 min at RT, rinsed with tap water until the excess dye was removed, and cleaned in 0.02% ammonia water until the tissue turned blue. Finally, the slides were completely dried at 60 °C, dipped in fresh xylene, and sealed with neutral gum.
Fluorescent in situ hybridization (FISH) was conducted using the RNAscope Fluorescent Multiplex kit (#323100, ACD) according to the manufacturer’s instructions. Mice were deeply anesthetized and perfused transcardially with 4% paraformaldehyde in PBS. Brains were dehydrated in 20% and 30% sucrose/PBS solutions for 36 h each and sectioned coronally at 15 μm thickness using a −20 °C cryostat (CM1850, Leica). The brain sections were treated with hydrogen peroxide for 10 min at RT, followed by target retrieval for 5 min at 99 °C, and protease treatment for 30 min at 40 °C. The Cdh11 probe was hybridized to the brain sections for 2 h at 40 °C. Signal amplification was performed by sequential application of multi-channel fluorescence Amp 1 (30 min at 40 °C), Amp 2 (30 min at 40 °C), and Amp 3 (15 min at 40 °C). TSA Plus fluorophore (1:750) was then applied to the sections and incubated for 30 min at 40 °C.
For FISH and immunofluorescence double labeling, hybridized sections were incubated overnight at 4 °C with the primary antibody against GAD67 (mouse, 1:250, #MAB5406, Millipore) or Neurogranin (rabbit, 1:1000, #ab217672, Abcam). This was followed by a 2-h incubation at RT with the secondary antibody (goat-anti-mouse IgG or goat-anti-rabbit IgG, 1:1000, conjugated with Alexa Fluor 488, #A-11029; #A-11034, Thermo Fisher). DAPI staining solution was added and incubated at RT for 30 seconds, followed by application of ProLong Gold Antifade mounting medium (P0126, Beyotime). A 24 mm × 50 mm glass coverslip was placed over the tissue sections. Images were captured using an FV10i confocal microscope at 60x magnification (Olympus, Japan).
For FISH after retrograde tracing, anesthetized mice were perfused transcardially 30 days following the injection of the retrograde tracing virus (AAV2/Retro-hSyn-EGFP-WPRE-pA) into the DS, NAc, or BLA using stereotactic injection method. Cryostat brain slices (15 μm thickness) were processed for the FISH procedure. Neurons that were retrogradely labeled were counterstained using a GFP antibody (rabbit, 1:500, #2956s, CST).
Stereotactic surgery
Mice anesthetized with 2% sodium pentobarbital in ddH2O (20 mg/kg) were secured in a stereotaxic injection frame (RWD Life Science Co., LTD, Shenzhen, China). Viral vectors were injected with a microinjection needle (Hamilton, 5 μL) at a speed of 40 nL/min using a micro-injection pump (TJ-4A, Longerpump, Hebei, China). 10 min after the injection completed, the needle was retracted at a speed of 0.5 mm/min. The animals were left undisturbed in their home cage for 3 weeks to recover from surgery and allow vector expression before behavioral tests. Needle placements and injection volumes were as follows: ACC (AP: +0.3 mm, ML: ±0.4 mm, DV: −1.75 mm, 200 nL), NAc (AP: +1.3 mm, ML: ±1.4 mm, DV: −4.6 mm, 200 nL), DS (AP: +1.3 mm, ML: ±1.4 mm, DV: −2.6 mm, 300 nL), BLA (AP: −1.46 mm, ML: ±2.9 mm, DV: −4.7 mm, 200 nL).
The viral vectors used and their respective titers were as follows: AAV2/9-CaMKIIα-EGFP-Cre-WPRE-pA (5.50 ×1012 vg/mL); AAV2/9-CaMKIIα-EGFP-WPRE-pA (5.60 ×1012 vg/mL); AAV2/Retro-hSyn-Cre-mCherry-WPRE-pA (5.50 ×1012 vg/mL); AAV2/Retro-hSyn-EGFP-WPRE-pA (5.60 ×1012vg/mL); AAV2/9-CaMKIIα-hM4Di-EGFP-WPRE-pA (5.16 ×1012 vg/mL); AAV2/9-CaMKIIα-DIO-hM4Di-mCherry-WPRE-pA (2.66 ×1012 vg/mL); AAV2/Retro-hSyn-Cre-EGFP-WPRE-pA (5.50 ×1012 vg/mL); AAV2/9-CaMKIIα-hM3Dq-EGFP-WPRE-pA (5.16 ×1012 vg/mL); AAV2/9-CaMKIIα-GCaMp6m-WPRE-pA (5.00 ×1012 vg/mL); AAV2/9-CaMKIIα-FDIO-hM3Dq-P2A-mCherry (5.00 ×1012 vg/mL); AAV2/retro-hsyn-Flp-P2A-EGFP (5.00 ×1012 vg/mL); AAV2/9-CaMKIIα-Cdh11-3×flag-WPRE-pA (5.30 ×1012 vg/mL). Virus vectors used in this study were purchased from BrainVTA Co., Ltd (Wuhan, China), BrainCase Co., Ltd (Wuhan, China) and OBiO Corp., Ltd (Shanghai, China).
Fiber photometry recording
Three weeks after injecting AAV2/9-CaMKIIα-GCaMp6m-WPRE-pA into the mouse ACC (AP: +0.3 mm, ML: ±0.4 mm, DV: −1.85 mm), an optical fiber (FC: 200 μm diameter, 2.5 mm length, NA 0.37, Shanghai Zhuen Bio-Technology Co., Ltd, China) was implanted (AP: +0.3 mm, ML: ±0.4 mm, DV: −1.75 mm). A week after optical fiber implantation, calcium transients were recorded using a three-color single-channel fiber optic recording system (Thinker Tech, Nanjing, China). Before recording, a laser power meter (Sanwa, Japan) was used to adjust the 405 nm light to 25–30 μW and the 470 nm light to 30–40 μW. The fiber jumper (RWD Life Science Co., LTD, Shenzhen, China) was placed in a dark environment to record the offset signal. It was then connected to the implanted optical fiber. Once the signal stabilized, the mouse was placed in a clean cage and allowed to move freely for 30 min while simultaneously recording the fluorescent signal and mouse behavior. Fluorescent signals were acquired at a 100 Hz sampling rate, with the 470 nm light exciting GCaMP6m fluorescence and the 405 nm channel serving as the control. The change in calcium signal before and after self-grooming was measured using the formula ΔF/F = (F - F0) / (F0 - offset), where F is the fluorescence signal, and F0 is the baseline fluorescence signal averaged over a 2-second window before self-grooming. The degree of calcium fluctuation was estimated by the peak-to-valley amplitude of ΔF/F% or by the ratio of coefficient of variation (CV%). The CV% is calculated as the standard deviation of F divided by the mean of F, multiplied by 100%. The heatmap and averaged calcium traces were plotted using Prism 10 (version 10.3.1, 509) or eGPS software (Big Data center, Beijing).
Chemogenetic manipulation
Behavior tests were conducted 3 weeks after injecting mice with inhibitory (hM4Di) or excitatory (hM3Dq) chemogenetic viral vectors. Clozapine N-oxide (CNO) was administered intraperitoneally using a 1 mL syringe at doses of 3.5 mg/kg for inhibition and 1.75 mg/kg for activation. The control groups received parallel injection of the same chemogenetic vectors as the experimental group but were treated with an equal volume of saline instead of CNO. The behavior test was performed 45 min after injection of CNO or saline.
Whole-cell patch clamp recording
Mice anesthetized with 2% sodium pentobarbital in ddH2O (20 mg/kg) were perfused intracardially with 10 mL ice-cold oxygenated solution A, containing 92 mM NMDG, 2.5 mM KCl, 30 mM NaHCO3, 1.25 mM NaH2PO4, 20 mM HEPES, 25 mM D-(+)-Glucose, 5 mM Na-ascorbate, 3 mM Na-pyruvate, 2 mM thiourea, 10 mM MgSO4 (liq), and 0.5 mM CaCl2 (liq). The mouse brain was quickly removed and placed into ice-cold oxygenated solution A. Coronal sections containing the ACC region (300 μm thickness) were cut with a vibratome (Leica VT1200S). The slices were incubated for 30 min at 34 °C in solution A and then incubated for 1 h at RT in solution B, containing 119 mM NaCl, 2.5 mM KCl, 24 mM NaHCO3, 1.25 mM NaH2PO4, 12.5 mM D-(+)-Glucose, 2 mM MgSO4 (liq), and 2 mM CaCl2 (liq). Both solution A and B were freshly prepared and saturated with a 95%:5% mixture of O2:CO2.
After incubation, the brain slice was transferred to the recording chamber, which was continuously perfused with solution B at a rate of 2 mL/min. Glass pipette with a resistance of 4–7 MΩ was used and contained a solution comprising 135 mM potassium gluconate, 7 mM KCl, 10 mM HEPES, 10 mM phosphocreatine, 4 mM ATP-Mg, and 0.4 mM GTP-Na, adjusted to a pH of 7.2–7.3 and an osmolarity of 285 mOsm. Under fluorescence microscope, fluorescent cells were identified. Positive pressure was applied to the glass pipette as it was brought close to the cell surface until small bubbles emerged. Subsequently, the positive pressure was removed to establish a high-resistance seal, and negative pressure was applied to rupture the membrane, thereby achieving the whole-cell recording mode.
Data was collected using a MultiClamp 700B amplifier (Molecular Devices, CA, USA), with a low-pass filter set at 3 kHz and a sampling frequency of 10 kHz. Action potentials of the cell were recorded in current-clamp mode, with the membrane voltage clamped at −60 mV, the duration of the test current was 500 ms, the frequency was 0.1 Hz, and the stimulus intensity was increased from 60 pA to 200 pA in steps of 20 pA. Cells exhibiting a series resistance exceeding 25 MΩ were excluded from the analysis. To test the effectiveness of chemogenetic inhibition, brain slices were perfused with solution B containing 5 μM CNO, and the number of action potentials discharged was recorded before, during, and after CNO treatment.
Synaptosome extraction
Cortical tissues (0.2 g) were collected in a 2 mL centrifuge tube and homogenized in 1.5 mL of ice-cold homogenization buffer. The buffer contained 0.32 M sucrose, 100 mM PMSF, 100 mM protease inhibitor Cocktail, 100 mM Na3VO4, 2 M DTT in a ratio of 1000:10:1:1: 1. After homogenization, the lysate was centrifuged at 1000 xg, 4 °C for 15 min. The pellet (P1) was discarded, and the supernatant (S1) was transferred to a 1.5 mL Eppendorf tube and centrifuged at 10000 xg for 15 min, 4 °C. This yielded a pellet (P2), representing the crude synaptosome fraction, and a supernatant (S2).
P2 was resuspended in 2 mL of 0.32 M sucrose and then transferred to a 5 mL tube. Next, 3 mL of 0.8 M sucrose was slowly injected into the bottom of tube using a syringe. The tube was gently chilled on ice and centrifuged at 3900 rpm for 18 min. After centrifugation, the flocculent white substances located in the middle layer of the tube were carefully extracted using a 1 mL syringe. This white substance was then transferred to a clean tube followed by centrifugating at 21000 xg, 4 °C for 45 min. The resulting pellets (P3), which contained synaptosome, was further processed in lysis buffer to carry out Western blotting analysis or used in preparation of pre- and post-synaptic components as follows:
P3 was suspended in 200 μL ice-cold HB buffer (0.32 M sucrose, 5 mM HEPES, pH 7.4), transferred to a 1.5 mL tube, and centrifuged at 10000 xg for 10 min. The resulting supernatant was discarded, and the pallet (P4) was fully resuspended in 500 μL RB buffer (0.32 M sucrose, 1 mM NaHCO3) and 500 μL LB buffer (1% TritonX-100, 12 mM Tris-HCL, pH 7.4). The mixture was transferred to a 1.5 mL tube and centrifuged at 33000 xg, 4 °C for 20 min. The resulting pellets contained post-synaptic components, while the supernatant contained the pre-synaptic components.
Protein sample preparations and immunoblotting assay
Target tissue was homogenized in a tube containing ceramic beads and lysis buffer (10% glycerol, 2% SDS, 1.25% β-mercaptoethanol, 25 mM Tris-HCl, pH 6.8, 12.5 mM EDTA, 1/50 tablet of cOmplete™ EDTA-free protease inhibitor cocktail) using a Precellys® 24 grinder (Bertin Technologies, France). The homogenate was boiled at 100 °C for 10 min and then centrifuged at 12,000 xg, 4 °C for 10 min. The protein supernatant was divided into small aliquots and stored at −80 °C. One aliquot was used to determine the protein concentration using the Quick Start™ Bradford protein assay kit (Bio-Rad, USA).
Protein samples were resolved via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene fluoride (PVDF) membrane and blocked with 5% milk in TBST for 2 h at RT. Blots were incubated with primary antibodies overnight at 4 °C and then washed 3 times for 10 min with TBST. The primary antibodies used were CDH11 (#321700, Thermo Fisher), MAGI2 (#25189-1-AP, Proteintech), VTI1A (#12354-1-AP, Proteintech), UNC13A (#126103, Synaptic Systems), SYT1 (#105011, Synaptic Systems), STX1B (#110403, Synaptic Systems), STX17 (#17815-1-AP, Proteintech), PSD95 (#MA1-045, Thermo Fisher), GLUR1 (#ab109450, Abcam), SYP (#MAB5258-1, Millipore), NR2A (#4205, CST), NR2B (#4207, CST), β-ACTIN (#66009-1-Ig, Proteintech), and GAPDH (#10494-1-AP, Proteintech). The blots were then incubated with HRP-conjugated secondary antibodies: goat-anti-mouse antibody (1:1000, #ab216772, Abcam) or goat anti-rabbit antibody (1:1000, #ab175773, Abcam) for 2 h at RT and washed with TBST. Protein bands on the blots were visualized using BeyoECL Moon kit (Beyotime, Shanghai, China). Images were captured with UVP Chemstudio touch (Analytik Jena, USA), and all protein bands detected were quantified using VisionWorks® 11.1 (Analytik Jena, USA). The relative quantity of a protein was determined by dividing the density value of its band by the density value of the GAPDH or β-actin band in the same lane on the blot. All images of the immunoblots and PVDF membranes used in this study are provided in the Supplementary WB file.
Immunoprecipitation
Immunoprecipitation (IP) was performed using the Dynabeads™ Protein G Immunoprecipitation Kit (#10007D, Thermo Fisher, USA). Cortical tissue from P7 mice was homogenized in a lysis buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40, 1 mM EDTA and protease inhibitors. A 10 μL anti-CDH11 (#4442, CST) antibody was diluted in 160 μL Ab Binding and Washing Buffer and incubated with 30 μL magnetic beads for 10 min at RT.
The magnetic bead-antibody complex was then resuspended in 200 μL Ab Binding and Washing Buffer and incubated with 200 μL of protein extract (15 μg/μL) for 2 h at RT. After removing the supernatant, the complex was washed 3 times using 200 μL Washing Buffer. For Western blotting, lysis buffer (10% glycerol, 2% SDS, 1.25% β-mercaptoethanol, 25 mM Tris-HCl, pH 6.8, 12.5 mM EDTA, 1/50 tablet of cOmplete™ EDTA-free protease inhibitor cocktail) was added to the complex, heated for 10 min at 70 °C, and the supernatant was collected for SDS-PAGE.
Golgi staining technique
The Golgi stain kit (#K401, FD Neuro Technologies, USA) was used following the manufacturer’s instructions. After cervical dislocation, the mouse brain was quickly removed, washed with ddH2O to remove blood, and soaked in an equal volume mixture of A/B solution. The brain was stored at RT for 10 days, then transferred to solution C and stored at RT for 3 days. The tissues were then frozen in liquid nitrogen and sectioned coronally at 120 μm thickness. After drying 2-3 days at RT, the slices were washed twice with ddH2O for 4 min each time. A mixture of D/E solution was used for 10 min of alkalization. After washing twice with ddH2O, the sample was dehydrated using ethanol diluents (50%, 75%, 95%, and 100%) for 4 min each. Xylene was used 3 times for clearing, each for 4 min. Finally, the slides were sealed with neutral resin. The entire staining process was conducted in the dark. Images were captured using Tissue Cytometry Analysis System (TissueGnostics GmbH, Austria).
Electron microscopy
Adult male mice were anesthetized with sodium pentobarbital and perfused with 4% paraformaldehyde. The brains were post-fixed in 4% paraformaldehyde at 4 °C for 24 h, then transferred to 2.5% glutaraldehyde for further fixation over 3 days. The tissues were washed 3 times with 0.1 M sodium carbonate for 10 min each time. After fixation with 1% osmium acid for 2 h, the tissues were washed 3 times with ddH2O under dark conditions, 10 min for each time. The tissues were mordanted with 2-thiophenecarboxylic acid hydrazide (TCH) at RT for 10 min, followed by 3 washes in ddH2O for 10 min each. Under dark conditions at 4 °C, the tissues were stained with 1% uranium acetate for 40 min and then washed 3 times with ddH2O for 10 min each time. Next, the tissues were incubated with lead aspartate at 60 °C for 30 min and washed 3 times with ddH2O for 10 min each time.
Gradient dehydration was performed using ethanol diluents (30%, 50%, 70%, 90%, and twice with 100%), with each step lasting 10 min. The samples were then treated with propylene oxide for 20 min and soaked in a mixture of propylene oxide and propylene oxide resin (1:1) at 4 °C overnight. Subsequently, the tissues were soaked in 100% epoxy resin 3 times, each soak lasting 2 h. The samples were sliced using a Leica ultra-slicer at a thickness of 65 nm, and images were captured using a transmission electron microscope (JEM2100, JEOL, Japan) at 5000x magnification.
Protein sample preparations and mass spectrometry analysis
Proteomic analysis of mouse ACC proteins was performed using a TMT-based mass spectrometry analysis by Biotree Biomedical Technology Co., LTD (Shanghai, China). Briefly, 0.01 g of ACC tissue was homogenized in 100 μL of ice-cold RIPA (25 mM Tris-HCl, pH 7.6, 15 mM NaCl, 1% NP40, 1% sodium deoxycholate, 1% SDS, and proteinase inhibitors) in a 1.5 mL Eppendorf tube by ultrasonication in an ice-bath. A completed homogenate was centrifuged at 12,000 rpm at 4 °C for 15 min. The resulting supernatant was transferred to a new 1.5 mL tube, and the protein concentration was determined using the Enhanced BCA Protein Assay Kit (#P0010, Beyotime, China) according to the manufacturer’s instructions.
To precipitate the proteins, the supernatant was mixed with five volumes of −20 °C acetone and kept at −20 °C overnight. The mixture was then centrifuged for 10 min at 12,000 rpm, 4 °C. The resulting pellet was collected and rinsed twice with 80% acetone. The precipitated proteins were dissolved in a solution containing 100 mM HEPES and 1% SDS using ultrasonication in a 1.5 mL Eppendorf tube. DTT was then added to a final concentration of 5 mM, and the mixture was incubated in a shaker at 55 °C for 20 min to reduce protein disulfide bonds. After cooling, iodoacetamide was added to the mixture to a final concentration of 15 mM and incubated in the dark for 30 min to alkylate the reduce protein disulfide bonds.
The alkylated proteins were digested overnight with 0.5 μg/μL sequence-grade modified trypsin (Promega, Madison, WI, USA) at an enzyme-to-substrate ratio of 1:50, 1000 rpm and 37 °C. Post-digestion, the peptide samples were labeled using the TMTsixplexTM Isobaric Label Reagent Set (Thermo Scientific, Rockford, USA) according to the manufacturer’s instructions. To remove sodium deoxycholate, trifluoroacetic acid (TFA) was added to the peptide solution to a final concentration of 2%. The mixture was centrifuged at high speed, and the supernatant was transferred to a new 1.5 mL tube. An additional 1 mL of 2% TFA was mixed with the supernatant and centrifuged at 12,000 rpm for 10 min to obtain the labeled peptide sample. Finally, the peptides were desalted using a C18 tip column (Changsha Huaxue Biotechnology Co., Ltd, Changsha, China), following the manufacturer’s instructions. The recovered peptides were then vacuum-dried at 4 °C.
The lyophilized peptide samples were reconstituted with 50 μL of mobile phase A (10 mM ammonium acetate aqueous solution, pH 10) and separated using high pH reversed-phase ultraperformance liquid chromatography under alkaline conditions. Chromatographic separation was performed on a Waters XBridge BEH C18 XP Column (150 mm × 2.1 mm, 2.5 μm, USA) using mobile phase B (10 mM ammonium acetate, 10% H2O, 90% acetonitrile, pH 10). The liquid phase gradient over 60 min was as follows: 5% mobile phase B, 2 min; 5% to 30% mobile phase B over 40 min; 30% to 40% mobile phase B over 10 min; 40% to 90% mobile phase B over 4 min; 90% mobile phase B, 2 min; 2% mobile phase B, 2 min. The fractions were collected every min, divided into 12 fractions, vacuum dried, and then frozen at −80 °C.
Approximately 2 µg of total peptides were separated and analyzed using a nano-UPLC system (EASYnLC1200) connected to a mass spectrometer (Q Exactive HFX) with a nanoliter ion source. Chromatographic separation was performed using an analytical reverse phase ReproSil-Pur 120 C18 AQ column (100 µm ID × 15 cm,1.9 µm, Dr. Maisch, Germany). The mobile phases used were phase A: 0.1% formic acid, 98% water, 2% acetonitrile and phase B: 0.1% formic acid, 80% acetonitrile, 20% water. The column was equilibrated with 100% phase A. The liquid phase gradient over 90 min, with a flow rate of 300 nL/min, was set as follows: 25% mobile phase B, 2 min; 5% to 22% mobile phase B over 68 min; 22% to 45% mobile phase B over 16 min; 45% to 95% mobile phase B, 2 min; 95% mobile phase B, 2 min.
Mass spectrometry analysis was performed over a total of 90 min using data-dependent acquisition (DDA) mode with positive ion detection. The MS1 acquisition range was 350-1600 m/z, with a resolution of 120 k at m/z 200. The automatic gain control (AGC) target value was set to 3E6, with a maximum ion injection time (max IT) of 30 ms. The top 20 most intense ions were selected for fragmentation with a quadrupole isolation window of 0.7 m/z and a normalized collision energy of 32%. The minimal MS2 acquisition range was set to 110 m/z with a resolution of 15 k. The AGC target value was set to 1E5, with a max IT of 96 ms. The dynamic exclusion time was 45 s, and single-charged ions and ions with a valence higher than 6 were excluded.
All raw MS files were processed using Proteome Discoverer (PD) software (Version 2.4.0.305) with the built-in Sequest HT search engine. MS spectra lists were searched against Uniport databases (Mus musculus 10090 2021 8.fasta). Fixed modifications included carbamidomethylation of cysteine (C), TMT 6-plex labeling at lysine (K), and N-termini, while variable modifications included oxidation of methionine (M) and acetylation of protein N-termini. Trypsin was the protease used and 2 missed cleavages were allowed. The false discovery rate (FDR) was set to 0.01 for both peptide-spectrum match and peptide levels. Peptide identification allowed an initial precursor mass deviation of up to 10 ppm and a fragment mass deviation of 0.02 Da. Unique and Razor peptides were used for protein quantification, with total peptide amount used for normalization. All other parameters were set to default.
Proteomics analysis of synaptic proteins isolated from mouse cortex were performed by Biotree Biomedical Technology Co., LTD (Shanghai, China) using a label-free quantification (LFQ) method. Briefly, the synaptic pellet (P3) was washed twice by 200 µL, 80% acetone, and the pellet was carefully collected in a 1.5 mL Eppendorf tube. The pellet was dissolved in a solution containing 100 mM HEPES and 1% SDS by ultrasonication for 5 min, and DTT was then added to a final concentration of 5 mM. The mixture was incubated in a shaker at 55 °C for 10 min to reduce protein disulfide bonds. After cooling, iodoacetamide was added to the mixture to a final concentration of 10 mM and incubated in the dark for 15 min to alkylate the reduce protein disulfide bonds.
Alkylated proteins were digested overnight with 0.5 μg/μL sequence-grade modified trypsin at an enzyme-to-substrate ratio of 1:50, 1000 rpm and 37 °C. Trifluoroacetic acid (TFA) was added to the peptide solution to a final concentration of 2%, pH < 2, to remove sodium deoxycholate. The mixture was centrifuged at high speed, and the supernatant was transferred to a new 1.5 mL tube. 100 μL of 2% TFA was mixed with the supernatant and centrifuged at 13,000 rpm for 10 min to obtain protein peptides. The peptides were desalted using a C18 tip column (Changsha Huaxue Biotechnology Co., Ltd, Changsha, China), following the manufacturer’s instructions. The recovered peptides were then vacuum-dried at 4 °C.
Approximately 1 µg of total peptides were separated and analyzed using a EASYnLC1200 coupled to a Q ExactiveTM HFX with the nanoelectrospray ion source. Chromatographic separation was performed using an analytical reverse phase ReproSil-Pur 120 C18 AQ column (100 µm ID × 15 cm,1.9 µm, Dr. Maisch, Germany). The mobile phases used were phase A: 0.1% formic acid, 98% water, 2% acetonitrile and phase B: 0.1% formic acid, 80% acetonitrile, 20% water. The column was equilibrated with 100% phase A. The liquid phase gradient over 120 min, with a flow rate of 300 nL/min, was set as follows: 2%-5% mobile phase B over 2 min; 5% to 22% mobile phase B over 88 min; 22% to 45% mobile phase B over 26 min; 45% to 95% mobile phase B, 2 min; 95% mobile phase B, 2 min.
Mass spectrometry analysis was performed over a total of 120 min using data-dependent acquisition (DDA) mode with positive ion detection. Full scan range for MS1 acquisition was 350-1600 m/z, with a resolution of 120k at m/z 200. The automatic gain control (AGC) target value was set to 3E6, with a maximum ion injection time (max IT) of 50 ms. The top 20 most intense ions were selected for fragmentation with a quadrupole isolation window of 1.2 m/z and a normalized collision energy of 27%. The MS2 acquisition was at a resolution of 15 k with a dynamic first mass. The AGC target value was set to 1E5, with a max IT of 110 ms. The dynamic exclusion time was 45 s, and single-charged ions and ions with a valence higher than 6 were excluded.
All raw MS files were processed using Proteome Discoverer (PD) software (version 2.4.0.305) with the built-in Sequest HT search engine. MS spectra lists were searched against Uniport databases (Mus musculus 100902020 10.fasta). Fixed modification was carbamidomethylation of cysteine (C), and the variable modifications were oxidation of methionine (M) and acetylation of protein N-termini. The protease used was trypsin and 2 missed cleavages were allowed. The false discovery rate (FDR) was set to 0.01 for peptide-spectrum match and peptide levels. Peptide identification allowed a precursor mass deviation of 10 ppm and a fragment mass deviation of 0.02 Da. Unique and Razor peptides were both used for protein quantification, with total peptide amount used for normalization. All other parameters were set to default.
Identification and quantification of proteins precipitated by CDH11 were carried out using 4D-SmartDIA quantitative proteomic analysis, provided by Jingjie PTM Biolabs Co. Ltd. (Hangzhou, China). Independent replicates of protein samples precipitated by CDH11 were collected from three Cdh11 KO mice and two wild-type littermates, resulting in a total of five samples. These samples were placed in separate 1.5 mL Eppendorf tubes. A 5 μL aliquot from each tube was adjusted to pH 8.0 using ammonium bicarbonate, subjected to SDS-PAGE, and subsequently visualized using a PAGE-Silver staining kit (Solarbio, Beijing, China). Relative protein concentration of each sample was determined by comparing the total band density in each lane to that of control protein samples from HeLa cells run on a separate lane of the same gel.
On-bead digestion of each sample was performed with 20 ng/μL trypsin (Promega, USA) overnight at 37 °C. Post-digestion, the sample was reduced in a solution containing 5 mM dithiothreitol for 1 h at 37 °C. Samples were then alkylated in a 11 mM iodoacetamide (Sigma-Aldrich, USA) for 45 min at room temperature in darkness. After alkylation, the samples were redigested with 10 ng/μL trypsin for 4 h at 37 °C. Finally, the digested samples were acidified 1:10 v/v with 10% TFA and desalted by the Strata X C18 SPE column (Phenomenex, USA).
A 0.25 μg aliquot of tryptic peptides was dissolved in liquid chromatography mobile phase A and then were separated using the Easy-nLC1000 ultra-high performance liquid chromatography (UHPLC) system. Mobile phase A comprised an aqueous solution with 0.1% formic acid and 2% acetonitrile, while phase B consisted of an acetonitrile-water solution with 0.1% formic acid. The liquid phase gradient was conducted as follows: 6%-24% B over 9 min; 24%-35% B over 2 min; 35%-90% B over 2 min; 90% B over 2 min. The flow rate was maintained at 500 nL/min.
After UHPLC separation, peptides were ionized using a capillary ion source and analyzed on an ion mobility spectrometry (TIMS) coupled with time-of-flight (TOF) Pro mass spectrometry (Bruker Daltonik, Bremen, Germany). The ion source voltage was set to 1.75 kV. Peptide parent ions and their fragments were detected and analyzed using TOF mass spectrometry. Data acquisition was performed in data-independent acquisition parallel accumulation-serial fragmentation (dia-PASEF) mode. The primary mass spectrometry (MS1) scan range was set to 300-1500 m/z. Following the acquisition of one MS1 spectrum, 20 PASEF mode acquisitions were performed. The secondary mass spectrometry (MS2) scan range was 400-850 m/z, with a window of 7 m/z for each acquisition.
DIA data were analyzed using the DIA-NN (v1.8) search engine with default parameters. The database used was Mus musculus 10090 SP 20231220.fasta, containing 17,191 sequences. The protease used was set to trypsin/P and 1 missed cleavage was allowed. Fixed modifications were N-terminal methionine excision and cysteine carbamidomethylation. A theoretical spectral library was constructed using a deep learning algorithm, and an anti-library was included to calculate the false discovery rate (FDR) resulting from random matches; the FDR for precursor identification was set to 1%.
The resulting data were plotted by HIPLOT (https://hiplot.cn/) and Wei Sheng Xin (https://www.bioinformatics.com.cn) online analysis tool.
Statistical analysis
Quantitative analyses were performed by an experimenter who was blinded to the experimental conditions whenever possible. Statistical analyses were conducted using Prism 10.3.1 (GraphPad Software Inc, USA). All data were first subjected to normal distribution detection using the Normality and Lognormality Test. Data that passed the normal distribution detection were examined by two-tailed Student’s t-test, while those that do not fit the normal distribution were examined by non-parametric Mann Whitney test. Statistical significance was set at P < 0.05. Randomization was not applied in this study, as experimental group assignment was based on predefined genotypes or treatment conditions. For representative images, similar results were obtained in at least two independent trials. Detailed information of statistical analysis and material resource for each experiment is shown in figure legends and in Supplementary Table 5.
Results
Expression of Cdh11 in ASD-related brain regions and circuits
We first examined the developmental expression profile of Cdh11 in mouse brains using in situ hybridization. We found that Cdh11 is highly expressed in key brain regions implicated in ASD pathogenesis, namely the anterior cingulate cortex (ACC), striatum, and hippocampus, at both embryonic day 17.5 (E17.5) and postnatal days 5 and 14 (P5, P14) (Fig. 1A). Additionally, immunofluorescent staining following fluorescence in situ hybridization (FISH) revealed intense Cdh11 signals within Neurogranin-positive pyramidal neurons in the ACC and hippocampus, as opposed to the minimal FISH signal within GAD67-positive GABAergic neurons (Fig. 1B and C). Conditional knockout of Cdh11 in the forebrain pyramidal lineage (EcKO), achieved by crossing the Cdh11-floxed allele with the Emx1-Cre line, significantly reduced the FISH signal in Neurogranin-positive cells in both the ACC and hippocampus (Fig. 1B and C). These results suggest a predominant expression of Cdh11 in glutamatergic excitatory neurons within these areas.
(A) In situ hybridization of Cdh11 at different developmental stages in mouse brains. ACC, anterior cingulate cortex; CPu, caudate putamen. (B and C) Fluorescence in situ hybridization (FISH) and immunofluorescence double staining indicate higher expression of Cdh11 in neurogranin-positive neurons compared to GAD67-positive neurons in the ACC (B) and hippocampus (C). (D) FISH following retrograde tracing indicates Cdh11 expression in ACC neurons that project to the nucleus accumbens (NAc), dorsal striatum (DS), and basolateral amygdala (BLA), and all scale bars are 200 µm. Both 3D projection images and the XZ and YZ optical planes illustrate the localization of FISH signal within retrogradely labeled neurons. See also Figure S1. (E and F) Representative images of Western blots and relative expression levels of CDH11 in the cortex (E) and hippocampus (F) at different developmental stages (n = 3 mice/stage). Relative expression levels of CDH11 are presented as mean ± S.E.M. A P value of ≤ 0.05 determined by Dunn’s nonparametric test was considered statistically significant. (G) Schematic of cortex protein isolation from synaptosomes (Syn), presynaptic membranes (Pre-Syn), and postsynaptic membranes (Post-Syn). (H and I) Western blotting of CDH11 in different synaptic fractions from wild-type (WT) and conditional knockout mice with Cdh11 gene ablation in the pyramidal lineage of the forebrain by the Emx1-Cre (EcKO). Red asterisks in (I) indicate the estimated molecular weight of CDH11. Cytosol, Cyt. See also Figure S2.
Considering the critical connectivity of the ACC to downstream brain regions, such as the dorsal striatum (DS), nucleus accumbens (NAc), and basolateral amygdala (BLA) (Supplementary Fig. 1), which are closely associated with ASD pathogenesis [26, 27], we conducted FISH analysis of Cdh11 expression after adeno-associated virus (AAV)-mediated retrograde tracing of ACC neurons that project to these brain regions. Thirty days post-injection of a retrograde tracing vector (AAV2/Retro-hSyn-EGFP) into one of these brain regions (DS, NAc, or BLA), FISH revealed Cdh11 mRNA within ACC neurons that project to either one of these downstream target regions (Fig. 1D).
We next analyzed the expression of CDH11 proteins in the cortex and hippocampus of mice at different developmental stages using Western blotting. A marked increase in CDH11 levels was observed starting from P7, suggesting its potential involvement in critical postnatal developmental processes, such as the synapse formation and dendritic spine pruning (Fig. 1E and F). To further validate the synaptic distribution of CDH11, we purified pre- and post-synaptic membrane fractions from cortical synaptosomes using the sucrose gradient centrifugation method [28] (Fig. 1G). Western blot analysis revealed the enrichment of CDH11 in synaptosomes and its presence in both pre- and post-synaptic membranes. CDH11 was absent from both the pre- and post-synaptic fraction following conditional knockout of Cdh11 in the forebrain pyramidal lineage (EcKO), consistent with its expression in forebrain pyramidal neurons (Fig. 1H and I).
Furthermore, the gene ontology (GO) analysis of genes co-expressed with CDH11 in developing human brain tissues identified a significant enrichment in molecular pathways associated with synaptic wiring. These pathways include “Regulation of synapse assembly,” “Positive regulation of axon extension,” and “Regulation of dendrite development,” along with the GO term “cell-cell adhesion” (Supplementary Fig. 2; Supplementary Table 1). Collectively, these findings indicate that CDH11, functioning as a homophilic adhesive molecule, may play a crucial role in the formation and maintenance of synaptic structures within brain regions and circuits implicated in ASD.
Increased self-grooming and reduced sociability of Cdh11-deficient mice
Using multiple behavioral assays, including the open field test, elevated plus maze test, modified three-chamber test, grip strength test, horizontal bar test, and rotarod test, we previously demonstrated that Cdh11-null mice exhibited core ASD-like behavioral deficits, such as significantly increased self-grooming and reduced sociability [17]. To confirm the impact of Cdh11 deficiency on ASD-related behavior and dissect the underlying molecular and circuit mechanisms, we employed the Cre/loxP recombination system to generate mice with Cdh11 gene ablation at defined developmental stages and specifically in a subpopulation of neurons. The loxP site was strategically inserted flanking the eighth exon of mouse Cdh11 gene (Fig. 2A). Subsequently, Cdh11-flox mice were crossbred with germline-specific Vasa-Cre [29], embryonic pyramidal lineage-specific Emx1-Cre [30], adult pyramidal neuron-specific CamKIIα-Cre [31], and embryonic interneuron-specific Dlx5/6-Cre mice [32]. This crossbreeding produced several mouse lines: general gene knockout (KO) mice, mice with forebrain pyramidal lineage-specific conditional knockout from early embryonic stages (EcKO), mice with forebrain pyramidal neuron-specific conditional knockout at adult (αcKO), and mice with cortical GABAergic neuron-specific conditional knockout from early embryonic stages (DcKO) (Fig. 2B). The significant reduction of CDH11 proteins in the cortex and hippocampus of KO, EcKO, and αcKO mice, but not the DcKO mice (Fig. 2C and D), is consistent with a high expression of CDH11 in the pyramidal lineage (Fig. 1B and C).
(A and B) Schematic representations of Cdh11-flox allele generation (A) and the cell-specific Cre expression periods in various mouse lines (B). (C and D) Western blots (C) and relative expression levels (D) of CDH11 in the ACC and hippocampus from KO, EcKO, αcKO, and DcKO mice and their WT littermates (n = WT/mutant: 3/3, 3/3, 6/6, and 3/3) (E–H) Quantification of the duration and frequency of self-grooming in KO (E), EcKO (F), αcKO (G) and DcKO (H) mice and WT littermates (n = WT/mutant: 14/13, 11/14, 12/10, and 11/15). (I–M) Schematic diagram (I) and quantitative results of the 3-stage sociability test in KO (J), EcKO (K), αcKO (L), and DcKO (M) mice and WT littermates (n = WT/mutant: 14/15, 20/13, 12/8, and 12/11). SI: Social interaction. Quantitative data from the Western blot analysis and the duration and frequency of self-grooming are presented as mean ± S.E.M., with individual values shown as dots. A P value of ≤ 0.05 determined by a two-tailed Student’s t-test was considered significant. Two-way ANOVA with Tukey’s multiple comparisons test was used for J to M. See also Figure S3.
Consistent with our previous findings in a Cdh11-null mouse line generated using a different transgenic strategy [17], the newly generated KO mice showed a significant increase in the duration of self-grooming (Fig. 2E) and abnormal social interactions in a three-stage sociability test (Fig. 2I and J). These results support an important role for CDH11 in the development and function of ASD-related brain circuits. EcKO mice also displayed significantly increased self-grooming and social deficits (Fig. 2F and K), traits not observed in αcKO and DcKO mice (Fig. 2G, H, L and M). This suggests that the repetitive behavior and social deficits caused by Cdh11 gene deficiency can be attributed to the loss of its developmental function in the pyramidal lineage of the forebrain. Moreover, we found that the water spray-induced grooming in KO and EcKO mice was largely unaffected compared to WT littermates (Supplementary Fig. 3A–C), suggesting that the peripheral sensory perception in KO and EcKO mice is normal and that the over-grooming after Cdh11 deficiency cannot be attributed to abnormal sensory perception.
In addition to the core symptoms of ASD, we found that EcKO mice showed a significant decrease in the fear response to a conditioned auditory cue, but not to the contextual cue, in the standard fear conditioning paradigm. In contrast, αcKO mice did not exhibit detectable abnormalities in the fear conditioning test (Supplementary Fig. 3D–F), supporting a developmental role of CDH11 (Figs. 1E, F, and 2). In the open field test, total locomotion and the duration of central exploration appeared normal for both EcKO and αcKO mice (Supplementary Fig. 3G–K).
To determine the critical period and brain regions in which Cdh11 deficiency leads to increased self-grooming, we employed virus-mediated Cre expression to specifically knock out Cdh11 in Cdh11-floxed mice. Given the high expression of Cdh11 in the developing ACC, we injected viral vectors (AAV2/9-CaMKIIα-Cre-EGFP) into the ACC of Cdh11-flox mice at P0, P7 and P30 to knock out Cdh11 specifically in the pyramidal lineage of this region (Fig. 3A–D), followed by behavioral tests in adulthood. We found that knockout of Cdh11 in ACC pyramidal neurons from P0 or P7 increased the duration and frequency of self-grooming, respectively (Fig. 3E–H), whereas ablation from P30 produced no detectable behavioral changes (Fig. 3I and J). In contrast, fear responses and social behavior were unaffected by Cdh11 ablation from P0 (Supplementary Fig. 4A–D).
(A) Schematic diagrams of CaMKIIα-Cre injections into the ACC of Cdh11-flox mice at P0, P7, and P30. (B–D) Western blot analysis of CDH11 expression in the ACC following injection of Cre-EGFP or control EGFP virus at P0 (B), P7 (C), and P30 (D) (n = EGFP/Cre: 4/4, 3/3, and 7/7). (E–J) Quantification of grooming duration (E, G, I) and frequency (F, H, J) in mice with or without Cdh11 knockout induced at P0, P7, or P30 via injection of Cre-EGFP or control EGFP virus (n = EGFP/Cre: 10/10, 7/6, and 9/8). (K) Schematic diagrams of AAV-CaMKIIα-Cdh11 injections into the ACC of adult EcKO and KO mice. (L) Western blot analysis of CDH11 expression in the ACC after virus injection on day 14, 21 and 30. (M–P) Quantification of grooming duration (M and O) and frequency (N and P) in EcKO (M and N) and KO (O and P) mice (n = 10/group). Data are presented as mean ± S.E.M., with individual values shown as dots. A P value of ≤ 0.05, determined by either a two-tailed or a one-tailed ($P) Student’s t-test or a two-tailed nonparametric Mann-Whitney test (nP), as appropriate, was considered significant. See also Figure S4.
To determine whether CDH11 restoration could reverse this phenotype, we injected AAV2/9-CaMKIIα-Cdh11-3xFlag into the ACC of 8-week-old KO and EcKO mice (Fig. 3K and L). Targeted expression of CDH11 in pyramidal neurons of the adult ACC significantly reduced the duration and frequency of self-grooming (Fig. 3M–P), indicating that CDH11 expression in this region is sufficient to mitigate the repetitive behavior. These results reinforce the important role of CDH11 in the development of ACC pyramidal neurons and emphasize the contribution of ACC circuits to the regulation of self-grooming.
Abnormal neuronal activities in the ACC of Cdh11-deficient mice
The expression of the immediate early gene c-Fos was used as an indicator to investigate neuronal activity in mutant mice during the observation of self-grooming (Fig. 4A). A marked reduction in c-Fos+ cells was observed in the ACC of EcKO mice compared to their WT littermates (Fig. 4B and C). No significant changes in c-Fos+ cells were observed in the BLA, DS, and NAc regions (Fig. 4D–H). These findings suggest a potential correlation between diminished neural activity in the ACC and the emergence of stereotypic behaviors in EcKO mice.
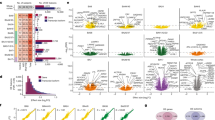
(A) Schematics of c-Fos staining followed by HomeCageScan behavioral observation. (B–H) Representative images and quantitative analyses of c-Fos+ cells in coronal sections of the ACC (B and C), DS and NAc (D–F), and BLA (G and H) from WT and EcKO mice (n = 5/group). (I and J) Schematics of the procedure used to record calcium signals in GCaMP-expressing neurons using fiber photometry. (K) Peri-event plots of calcium signals (ΔF/F%) of ACC pyramidal neurons before and during self-grooming. The solid curve and the shaded regions indicate the mean ± S.E.M. (L and M) Heat maps of the calcium fluctuations (ΔF/F%) of ACC pyramidal neurons before and during self-grooming in WT (L) and EcKO (M) mice. (N) The average coefficient of variation, CV (%), (O) peak-to-valley amplitude (ΔF/F%), and (P) total number of peaks during the pre-grooming period. (Q–S) Analyses of calcium signals during grooming of the mice: the highest value (Q) and lowest value (R) of ΔF/F% and the normalized area under curve (AUC) (S) (n = 30 trails from 6 WT mice, 45 trails from 5 EcKO mice). Quantitative data are presented as mean ± S.E.M. A P value of ≤ 0.05, determined by either a two-tailed Student’s t-test or a two-tailed nonparametric Mann-Whitney test (nP ≤ 0.05), as appropriate, was considered significant.
The calcium fiber photometry was further used to monitor the activity of pyramidal neurons within the ACC before and during self-grooming episodes in WT mice (Fig. 4I and J). A pronounced decline in the calcium signal was noted at the onset of self-grooming, indicating a significant association between neural activity dynamics in the ACC and the initiation of self-grooming behavior (Fig. 4K). We compared the calcium profiles in the ACC between WT and EcKO mice, recording the periods extending 30 seconds before and 10 seconds after the onset of self-grooming (Fig. 4L and M). During the pre-grooming phase, we calculated the coefficient of variation (CV) of each trial to reflect the amplitude of neuronal activity fluctuations. We found that the CV values in EcKO mice were significantly lower than those of WT littermates (Fig. 4N). Additionally, the calculation of the peak-to-valley amplitude (PV) in calcium signals of each trial showed that the PV values in EcKO mice were considerably lower than those in WT mice (Fig. 4O). However, the frequency of calcium transient peaks during this interval did not differ between the two groups (Fig. 4P). A decrease in the calcium signal was observed at the start of self-grooming in EcKO mice (Fig. 4K). However, the reduction in calcium signal amplitude in EcKO mice was significantly less than that in WT mice (Fig. 4R). These results collectively suggest that there is a link between the dynamics of neuronal activity in the ACC and the duration of self-grooming, and that the absence of the Cdh11 gene in pyramidal neurons attenuates neuronal network activity in the ACC, potentially enhancing the manifestation of stereotypic behaviors in mice.
Chemogenetic manipulation of ACC activities alters self-grooming
We inhibited pyramidal neuron activity in the ACC of WT mice using inhibitory DREADD construct hM4Di (human muscarinic M4) activated by clozapine N-oxide (CNO), delivered via virus injection (Fig. 5A). Successful inhibition was confirmed by reduced c-Fos+ cell counts (Fig. 5B–D) and by suppressed neuronal firing in hM4Di-expressing neurons in slice recordings (Fig. 5E–G). Following CNO treatment, mice exhibited a significant increase in duration of self-grooming (Fig. 5H and I), indicating that reduced ACC pyramidal neuron activity promotes repetitive behavior. Conversely, chemogenetic activation of ACC pyramidal neurons using the excitatory DREADD hM3Dq significantly reduced grooming behavior upon CNO treatment in WT, EcKO, and KO mice (Fig. 5S–U).
(A) Schematic illustration of infecting ACC pyramidal neurons by the viral vector hM4Di. (B–D) Validation of the effectiveness of chemogenetic inhibition in brain slices from virus vector-infected mice treated without (B) or with (C) CNO and quantification of c-Fos+ cells (D) (n = 12 slices from 2 WT/18 slices from 3 EcKO mice); (E–G) the effectiveness of chemogenetic inhibition is also validated by whole-cell patch clamp recording before, during, and after washing out of CNO treatment (E), showing the number of action potentials, spike counts (F), and the minimal injected current required to induce action potentials (G) (n = 11 cells from 3 mice/group). (H and I) The duration of grooming (H) and frequency of grooming (I) are shown (n = 9 mice/group). (J–L) Schematics of chemogenetic inhibition of neural circuits, ACC to NAc (J), DS (K), and BLA (L), respectively. (M–R) Grooming duration (M, O, Q) and frequency (N, P, R) after inhibition of ACC to NAc (n = 10 mice/group), ACC to DS (n = 9 mice/group), and ACC to BLA (n = 10 mice/group). (S) Schematic diagram of infecting ACC pyramidal neurons with viral vector hM3Dq and behavioral tests after virus vector infection. (T–U) Grooming duration after CNO and saline treatment in the same mice (T: n = 6 WT, 8 EcKO; U: n = 5 WT, 4 KO). (V) Schematics of chemogenetic activation of ACC to NAc. (W–Y) Grooming behavior in the same WT (W), EcKO (X), or KO (Y) mice after saline and CNO treatment (n = 5 mice/group). Data are presented as mean ± S.E.M, with individual values or brain slices shown as dots. A P value of ≤ 0.05, determined by either a two-tailed or a one-tailed ($P) Student’s t-test, was considered significant. Two-way ANOVA with Tukey’s multiple comparisons test was used for F. Seconds, s. See also Figure S5.
Cdh11 is expressed in ACC pyramidal neurons that project to multiple downstream targets, including the NAc, DS, and BLA. We therefore examined the behavioral consequence of selectively inhibiting ACC neurons projecting to each of these regions (Fig. 5J–L). Inhibition of ACC-NAc projections led to a significant increase in both grooming duration and frequency, whereas inhibition of ACC-DS and ACC-BLA projections resulted in elevated grooming duration only (Fig. 5M–R).
To investigate whether downstream regions of the ACC are affected in EcKO mice, we analyzed the expression of several synaptic proteins in the NAc, DS, and BLA. Western blot analysis revealed that protein levels of glutamate receptor 2 A (NR2A), postsynaptic density protein 95 (PSD95), and synaptophysin were significantly reduced in the NAc of EcKO mice (Supplementary Fig. 5A and B), while no substantial changes were observed in the DS or BLA (Supplementary Fig. 5C–F). Consistent with these molecular alterations, transmission electron microscopy showed widened synaptic clefts and increased presynaptic vesicle density in excitatory synapses of the NAc in EcKO mice compared to WT controls (Supplementary Fig. 5G–I). To assess the functional relevance of this projection, we selectively activated ACC-NAc neurons in WT, EcKO, and KO mice and found a significant reduction in both grooming duration and frequency across all genotypes (Fig. 5V–Y).
Altogether, these results demonstrate that the over-grooming phenotype in EcKO and KO mice is driven by reduced activity in ACC pyramidal neurons, and that the NAc is a key downstream target affected by altered ACC input.
Multi-layered proteomics reveals critical roles of CDH11 in synaptic vesicle recycling
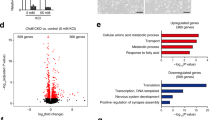
To elucidate the molecular mechanisms underlying decreased ACC activity due to Cdh11 deficiency, we first compared the proteomic profiles of the ACC between adult EcKO mice and their WT littermates. A total of 286 differentially expressed proteins were identified, with 159 downregulated and 127 upregulated in EcKO mice compared to WT littermates (Fig. 6A and B, Supplementary Fig. 6A–C; Supplementary Table 2). We conducted gene ontology (GO) analysis of downregulated proteins in the ACC of EcKO mice. Interestingly, the downregulated proteins were significantly enriched not only in expected cellular components and biological processes such as “Cell junction,” “Glutamatergic synapse,” and “Regulation of neuron projection development,” but also, unexpectedly, in components and processes like “Vesicle,” “Vesicle membrane,” and “Vesicle-mediated transport.” (Fig. 6A). GO analysis of upregulated proteins in the ACC of EcKO mice also highlighted molecular pathways closely related to “Vesicles,” “Synapse,” and “Endosome membrane” (Supplementary Fig. 6A). These results reinforce the findings of the GO analysis of CDH11 co-expressing genes in developing human brains, which also indicated a significant enrichment of these genes in the “Vesicle mediated transport” pathway (Supplementary Fig. 2). Consistent with the findings from proteomic profiling, Western blotting confirmed a significant reduction in the membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (MAGI2) and an increasing trend of the vesicle transport through interaction with t-SNAREs homolog 1 A (VTI1A) in the ACC of EcKO mice (Fig. 6C and D). MAGI2 functions as a scaffold molecule at synaptic junctions by assembling neurotransmitter receptors and cell adhesion proteins, while VTI1A mediates vesicle transport pathways through interactions with t-SNAREs on the target membrane [33, 34]. These results suggest a potential role for CDH11 in synaptic vesicle recycling in cortical glutamatergic synapses.
(A) Gene Ontology (GO) analysis for Biological Process (BP) and Cellular Component (CC) of downregulated proteins in the ACC of EcKO vs. WT mice. GO terms with a false discovery rate (FDR) of < 0.05. (B) Heatmap of differentially expressed proteins in the ACC of EcKO vs. WT mice. (C and D) Western blots (C) and relative expression levels (D) of CDH11, VTI1A, and MAGI2 in ACC (n = 6 WT/6 EcKO for CDH11 and VTI1A; n = 7 WT/7 EcKO for MAGI2). (E) GO analysis for BP and CC of downregulated proteins in cortical synaptosomes of EcKO vs. WT mice. GO terms with an FDR < 0.05. (F) Heatmap of differentially expressed proteins in cortical synaptosomes of EcKO vs. WT mice. (G and H) Western blots (G) and the relative expression levels (H) of CDH11, UNC13A, SYT1, STX1B (n = 7 mice/group) in cortical synaptosome preparations. (I and J) GO analysis (I) and volcano plot (J) of proteins immunoprecipitated by CDH11 from P7 cortical tissue lysates. (K) Schematic illustration of CDH11 co-immunoprecipitation. (L) Western blots of co-immunoprecipitated STX17 with CDH11. See also Figures S6-S8. (M and N) Representative images of excitatory (M) and inhibitory (N) synapses captured by transmission electron microscopy. (O–Q and R–T) Quantification of excitatory (O–Q) and inhibitory (R–T) synaptic ultrastructure: number of presynaptic vesicles (O and R), synaptic cleft width (P and S), and distance of synaptic vesicles to the presynaptic membrane (Q and T) (n = 3 mice/group). Data are presented as mean ± S.E.M, with individual values represented as dots. A P value of ≤ 0.05, determined by either a one-tailed ($P) or two-tailed Student’s t-test, or a one-tailed (n$P) or two-tailed (nP) nonparametric Mann-Whitney test, as appropriate, was considered significant.
To further investigate the potential role of CDH11 in synapse, we compared the proteomic profiles of cortical synaptosomes between adult EcKO mice and their WT littermates. A total of 354 differentially expressed proteins were identified, with 232 downregulated and 122 upregulated in EcKO mice compared to WT mice (Fig. 6E and F, and Supplementary Fig 6D–F; Supplementary Table 3). GO analysis indicated that downregulated synaptosome proteins in EcKO mice are enriched in biological processes “Synaptic vesicle cycle,” “Synaptic vesicle priming,” “Synaptic vesicle exocytosis,” “Cell adhesion,” and “Positive regulation of synapse assembly” (Fig. 6E). GO analysis of upregulated synaptosome components in EcKO mice also highlighted molecular pathways closely related to “Vesicles” and “Synapse” (Supplementary Fig. 6D), being similar to finding of the ACC proteomic analysis. Among those downregulated proteins, 16.4% (38/232) are located at glutamatergic synapses and 3.8% (9/232) at GABAergic synapses (Fig. 6E, Supplementary Table 3). The reduced expression of three proteins related to synaptic vesicle release in synaptosomes from EcKO mice, Protein unc-13 homolog A (UNC13A), Synaptotagmin-1 (SYT1), and Syntaxin-1B (STX1B), were confirmed by Western blotting (Fig. 6G and H). These results support an important role of CDH11 in the regulation of synaptic vesicle dynamics mainly in glutamatergic synapse.
We further examined proteins that interact with CDH11 during cortical development by profiling those co-immunoprecipitated with CDH11 in cortical tissues from neonatal (P7) WT mice, using cortical tissues from Cdh11 KO mice as the negative control (Fig. 6I–K; Supplementary Table 4). A total of 50 CDH11-interacting proteins were identified in the P7 cortex (Fig. 6J; Supplementary Table 4). GO analysis of these CDH11-interacting proteins revealed enrichment in molecular pathways associated with vesicle trafficking, including “Vesicle-mediated transport,” “ER to Golgi vesicle-mediated transport,” and “SNARE complex” (Fig. 6I). Western blot analysis was employed to confirm the interactions between CDH11 and Syntaxin-17, a key protein in membrane fusion (STX17) (Fig. 6L).
Together, multi-layered proteomic analyses suggest that CDH11 plays a critical role in vesicle trafficking at glutamatergic synapses during the postnatal development of the cerebral cortex.
Cdh11 deficiency decreases presynaptic vesicle density at excitatory synapses and increases the cleft width at inhibitory synapses
We found that Cdh11 deficiency has no effects on the cortical lamination (Supplementary Fig. 7) and has minimal effects on the dendrite development and dendritic spine formation for either apical dendrite or basal dendrite of ACC pyramidal neurons (Supplementary Fig. 8). Considering that Cdh11 deficiency significantly impacted synaptic components (Fig. 6), we further examined the ultrastructure of both excitatory and inhibitory synapses in the ACC of EcKO mice and their WT littermates using transmission electron microscopy. We observed a significant reduction in the number of presynaptic vesicles at excitatory synapses (Fig. 6M and O), characterized by a thick, dense postsynaptic density (asymmetric synapses), but not at inhibitory synapses (Fig. 6N and R), which have a thinner postsynaptic density (symmetric synapses), in the ACC of EcKO mice compared to WT mice. Although the distance between vesicles and the presynaptic membrane were largely unchanged (Fig. 6Q and T), the synaptic cleft width of inhibitory synapses (Fig. 6S), but not excitatory synapse (Fig. 6P), was significantly increased. Overall, these findings highlight the essential role of CDH11 in synaptic vesicle recycling within the mammalian forebrain.
Discussion
The present study reveals the critical role of CDH11 in synaptic development within key neuronal population associated with ASD-related brain circuits. Using multiple gene knockout and conditional knockout mouse models, we uncovered molecular and cellular mechanisms through which CDH11 influences the repetitive behaviors associated with ASD.
Expression and role of Cdh11 in the development of ASD-related circuit
We first demonstrated that Cdh11 is highly expressed in key brain regions implicated in ASD, including the ACC, striatum, and hippocampus, across various developmental stages. Notably, Cdh11 expression was predominantly observed in Neurogranin-positive pyramidal neurons. The conditional knockout of Cdh11 in the forebrain pyramidal lineage (EcKO) led to a significant reduction in CDH11 expression, confirming its neuronal specificity. Previous study [11] and multiple publicly available datasets of single-cell transcriptomes show that Cdh11 is also expressed in other cell types in the brain, including interneurons, endothelial cells, microglial cells, and Purkinje cells. However, our results show that the most abundant expression of Cdh11 occurs in pyramidal neurons, as its protein level was dramatically reduced in the cortex of EcKO mice. Together with the evidence that EcKO mice display core behavioral deficits of ASD, these results suggest the autistic phenotype caused by Cdh11 deficiency may be attributed primarily to its role in the development of forebrain pyramidal neurons.
Cadherin family members play crucial roles in multiple developmental processes, including cell proliferation, polarization, neuronal migration, axon projection, dendrite arborization, and synapse assembly, by mediating homophilic or heterophilic cell-cell interactions [35,36,37]. A previous study showed that Cdh11 expression increases in cells exiting the ventricular zone during embryonic development of the cerebral cortex. Overexpression of CDH11 in cortical progenitors via in-utero electroporation caused the premature exit of progenitors from the neuroepithelium and increased cell migration [38]. However, our previous work in Cdh11 KO mice [11] and the present work in EcKO mice did not show any apparent deficits in cortical lamination (Supplementary Fig. 7), suggesting that CDH11 may not play a major role in the proper radial migration or lamination of cortical pyramidal neurons. The lack of a prominent neuronal radial migration deficit could be attributed to the relatively low expression of CDH11 before P7 (Fig. 1E and F). Our finding of a significant increase in CDH11 expression after P7 is consistent with a previous report [18]. It suggests an important role for this adhesion molecule in postnatal development, particularly in synapse assembly, as supported by our proteomics and electron microscopy studies. Our findings suggest that synaptic deficits are the major cause of the behavioral deficits observed in Cdh11-deficient mice. However, a potential role for CDH11 in axon growth and pathfinding during development cannot be ruled out.
Behavioral implications of Cdh11 deficiency
Behavioral analyses revealed that both global and forebrain pyramidal lineage-specific knockout of Cdh11 in male mice caused over-grooming and social deficits, two core symptoms of ASD. These phenotypes were not observed in mice with adult-specific or GABAergic lineage-specific knockouts, suggesting that CDH11 functions specifically in pyramidal neurons during a critical developmental period. In addition to these core symptoms of ASD, we also found that the fear levels of EcKO mice were abnormally reduced, consistent with a previous study [25]. Abnormal fear responses have also been observed in several other ASD mouse models [39, 40]. The presence of multiple core and comorbid symptoms in Cdh11-deficient mice further supports the notion that Cdh11 is an important risk gene for ASD, and these mice could serve as an important animal model for investigating the neural mechanism of ASD-related behavioral defects. Furthermore, targeted ablation of Cdh11 in ACC pyramidal neurons from P0 or P7 replicated the over-grooming phenotype, whereas ablation at later postnatal stages did not, highlighting the critical role of CDH11 expression during development in the assembly of ASD-related circuits.
Although ASD is caused by developmental deficits, previous studies showed that restoring several ASD-risk genes like Shank3 [41], MECP2 [42], Ube3a [43], Nlgn3 [44], and SCN2A [45] at adolescent or adult can rescue autistic behavior in mouse models. Notably, we found targeted expression CDH11 in ACC of 8-week-old EcKO and KO mice effectively mitigated their over-grooming behavior. This result suggests that targeting CDH11 during adulthood through systemic gene therapy may hold potential for alleviating other ASD-associated symptoms, an intriguing possibility that deserves to be further explored in future studies.
The ACC is a high-level regulatory center in the brain, playing an important role in basic cognitive processes such as emotion, decision-making, learning, and motivation [46]. Functional abnormalities in the ACC are closely associated with stereotyped behaviors and abnormal fear and anxiety [47,48,49]. Pyramidal neurons in the ACC have extensive mutual connections with other brain regions, forming a complex neural network. Our findings provide substantial evidence that CDH11 is integral to the development and function of ACC pyramidal neurons, with its deficiency leading to significant neurodevelopmental and behavioral abnormalities characteristic of ASD.
We found that ablation of Cdh11 in ACC pyramidal neurons from P0 induced over-grooming but did not result in social deficits or abnormal fear responses. These findings suggest that the ACC primarily regulates repetitive behaviors, whereas other regions such as the hippocampus [50] and BLA [48] may contribute to social and emotional abnormalities, respectively. Whether CDH11 plays a similar role in these areas remains to be determined. Moreover, as all behavioral analyses in this study were conducted in male mice, future work should examine potential sex-dependent effects of Cdh11 deficiency.
Neuronal activity and circuitry alterations
We observed a marked reduction in c-Fos+ cells and attenuated calcium fluctuations in the ACC of adult EcKO mice, indicating diminished neuronal network activity. A previous study found that the calcium activity of neurons in dmPFC projecting to DS decreased significantly at the onset of self-grooming in mice [51]. In our study, we obtained similar results. We found that the calcium activity in the ACC of both WT and EcKO mice decreased significantly at the beginning of self-grooming, but the fluctuations of calcium activity in EcKO mice were significantly smaller than that in WT mice. Our electron microscopy also revealed a significant reduction in synaptic vesicles in glutamatergic synapses in the ACC of EcKO mice, which likely contributes to the reduced network activity. Consistent with these findings, a previous study in cultured hippocampal neurons showed that neurons from Cdh11 KO mice exhibit a significant reduction in neuronal activity and synchrony compared with WT neurons, suggesting that synaptic input is reduced in KO mice [25].
In addition to abnormalities in the presynaptic assembly, our results do not rule out the contribution of postsynaptic alterations to the reduced neuronal activity of ACC neurons. Our proteomics studies indicate changes in the expression of postsynaptic components including Gamma-aminobutyric acid type A receptor subunit alpha3 (GABRA3) and Ephrin type-A receptor 4 (EPHA4) and we found that the cleft width of inhibitory synapses was increased while the abnormality was not observed in excitatory synapses. Considering that inhibitory synapses in ACC pyramidal neurons of EcKO mice lack CDH11 only in the postsynaptic terminal, while excitatory synapses lack CDH11 on both the pre- and postsynaptic sides, it is plausible that unilateral knockout of CDH11 in the postsynaptic terminal of inhibitory synapses may lead to a significant increase in synaptic cleft width due to deficits in CDH11-mediated synaptic adhesion; In contrast, bilateral knockout may have stimulated compensatory mechanisms involving the upregulation of other adhesion molecules on both the pre- and postsynaptic sides to maintain the synaptic cleft width in excitatory synapses. Our results highlight the important role of CDH11 in excitatory synaptic input onto ACC pyramidal neurons. Future research investigating the role of CDH11 in the development and function of the output synapse of ACC pyramidal neurons and its impact on social and repetitive behaviors will enhance our understanding of its comprehensive functions in brain development and disease.
Molecular mechanisms underlying presynaptic function of CDH11
Consistent with the reduced neuronal activity in the ACC of EcKO mice, we found deficits in the ultrastructure of glutamatergic synapses in their ACC. CDH11 protein is enriched in synaptosomes, distributing at both pre- and post-synaptic membranes. Differentially expressed proteins in the cortex of EcKO mice were significantly enriched in pathways related to synaptic vesicle trafficking, suggesting a role for CDH11 in synaptic vesicle dynamics. Key CDH11-interacting proteins are involved in vesicle-mediated transport, reinforcing its role in synaptic vesicle trafficking. Additionally, ultrastructural analysis via electron microscopy showed a significant reduction in presynaptic vesicles at excitatory synapses, directly linking CDH11 deficiency to impaired synaptic vesicle trafficking and suggesting its pivotal role in the proper functioning of synaptic transmission and plasticity. Consistent with this notion, a previous study showed that hippocampal brain slices from Cdh11-null mice exhibit significantly lower LTP saturation levels [25]. These results strongly indicate that the absence of CDH11 can lead to deficits in vesicle circulation in glutamatergic synapse, thereby affecting the activity of ACC neuronal network.
Previous studies have shown that CDH2 (N-cadherin) interacts with Neuroligin-1 (NLGN1) to trans-synoptically enhances the recycling and clustering of presynaptic vesicles, at least in part by promoting the endocytosis of synaptic vesicles [52,53,54]. Interestingly, we found a significant downregulation of NLGN3 in synaptosomes from EcKO mice (Supplementary Table 3). Moreover, we identified the autophagosomal SNARE protein, STX17, among the CDH11-interacting proteins (Supplementary Table 4). Since STX17 interacts with SNAP29 and the endosomal/lysosomal SNARE VAMP8 [55], which are critical in regulating vesicle fusion and endocytosis, it is likely that CDH11 regulates the assembly of presynaptic vesicles in ACC pyramidal neurons through molecular mechanisms similar to those of CDH2. Our findings suggest that deficits in synaptic vesicle trafficking are a crucial cellular mechanism underlying the circuit and behavioral deficits observed in ASD and could be a potential intervention target for ASD.
In summary, we discovered that CDH11 plays an important role in the development and function of brain circuits associated with ASD, and its deficiency leads to significant behavioral deficits and synaptic abnormalities, potentially due to its important role in vesicle trafficking during synaptogenesis. Understanding the molecular mechanisms by which CDH11 defects contribute to core ASD symptoms will provide valuable insights into potential strategies to correct this deficit.
Data availability
Proteomics data are available via ProteomeXchange with identifier PXD055834 for ACC proteome, PXD056074 for cortex synaptosome proteome, PXD056411 for CDH11 co-immunoprecipitation proteome. This study did not generate new unique reagents. There are no restrictions to the availability of any reagent in this study. Supplementary information is available at MP’s website.
References
Courchesne E, Pierce K, Schumann CM, Redcay E, Buckwalter JA, Kennedy DP, et al. Mapping early brain development in autism. Neuron. 2007;56:399–413.
Zhou X, Feliciano P, Shu C, Wang T, Astrovskaya I, Hall JB, et al. Integrating de novo and inherited variants in 42,607 autism cases identifies mutations in new moderate-risk genes. Nat Genet. 2022;54:1305–19.
Fu JM, Satterstrom FK, Peng M, Brand H, Collins RL, Dong S, et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat Genet. 2022;54:1320–31.
Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–584.e23.
Dias CM, Walsh CA. Recent advances in understanding the genetic architecture of autism. Annu Rev Genomics Hum Genet. 2020;21:289–304.
Schmitz-Abe K, Sanchez-Schmitz G, Doan RN, Hill RS, Chahrour MH, Mehta BK, et al. Homozygous deletions implicate non-coding epigenetic marks in Autism spectrum disorder. Sci Rep. 2020;10:14045.
Ding C, Zhou W, Shi Y, Shan S, Yuan Y, Zhang Y, et al. Srcap haploinsufficiency induced autistic-like behaviors in mice through disruption of Satb2 expression. Cell Rep. 2024;43:114231.
Shih PY, Fang YL, Shankar S, Lee SP, Hu HT, Chen H, et al. Phase separation and zinc-induced transition modulate synaptic distribution and association of autism-linked CTTNBP2 and SHANK3. Nat Commun. 2022;13:2664.
Basu R, Duan X, Taylor MR, Martin EA, Muralidhar S, Wang Y, et al. Heterophilic Type II cadherins are required for high magnitude synaptic potentiation in the hippocampus. Neuron. 2018;98:658–68.
Bennison SA, Blazejewski SM, Liu X, Hacohen-Kleiman G, Sragovich S, Zoidou S, et al. The cytoplasmic localization of ADNP through 14-3-3 promotes sex-dependent neuronal morphogenesis, cortical connectivity, and calcium signaling. Mol Psychiatry. 2023;28:1946–59.
Wang C, Pan YH, Wang Y, Blatt G, Yuan XB. Segregated expressions of autism risk genes Cdh11 and Cdh9 in autism-relevant regions of developing cerebellum. Mol Brain. 2019;12:40.
Sato M, Nakai N, Fujima S, Choe KY, Takumi T. Social circuits and their dysfunction in autism spectrum disorder. Mol Psychiatry. 2023;28:3194–206.
Kelly E, Meng F, Fujita H, Morgado F, Kazemi Y, Rice LC, et al. Regulation of autism-relevant behaviors by cerebellar-prefrontal cortical circuits. Nat Neurosci. 2020;23:1102–10.
Munz M, Bharioke A, Kosche G, Moreno-Juan V, Brignall A, Rodrigues TM, et al. Pyramidal neurons form active, transient, multilayered circuits perturbed by autism-associated mutations at the inception of neocortex. Cell. 2023;186:1930–1949.e31.
Sindi IA. Implications of cell adhesion molecules in Autism Spectrum Disorder pathogenesis. J Microsc Ultrastruct. 2022;11:199–205.
Arutiunian V, Santhosh M, Neuhaus E, Sullivan CAW, Bernier RA, Bookheimer SY, et al. A common genetic variant in the Neurexin family member CNTNAP2 is related to language but not communication skills in youth with Autism Spectrum Disorder. Autism Res. 2025;18:898–908.
Wu N, Wang Y, Jia JY, Pan YH, Yuan XB. Association of CDH11 with Autism Spectrum Disorder revealed by matched-gene co-expression analysis and mouse behavioral studies. Neurosci Bull. 2022;38:29–46.
Frei JA, Niescier RF, Bridi MS, Durens M, Nestor JE, Kilander MBC, et al. Regulation of neural circuit development by Cadherin-11 provides implications for Autism. eNeuro. 2021;8:ENEURO.0066-21.2021.
Crepel A, De Wolf V, Brison N, Ceulemans B, Walleghem D, Peuteman G, et al. Association of CDH11 with non-syndromic ASD. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:391–8.
Costa CIS, da Silva Montenegro EM, Zarrei M, de Sá Moreira E, Silva IMW, de Oliveira Scliar M, et al. Copy number variations in a Brazilian cohort with autism spectrum disorders highlight the contribution of cell adhesion genes. Clinical Genetics. 2022;101:134–41.
Redies C, Hertel N, Hübner CA. Cadherins and neuropsychiatric disorders. Brain Res. 2012;1470:130–44.
Hawi Z, Tong J, Dark C, Yates H, Johnson B, Bellgrove MA. The role of cadherin genes in five major psychiatric disorders: a literature update. Am J Med Genet B Neuropsychiatr Genet. 2018;177:168–80.
Chen X, Xiang H, Yu S, Lu Y, Wu T. Research progress in the role and mechanism of Cadherin-11 in different diseases. J Cancer. 2021;12:1190–9.
Paradis S, Harrar DB, Lin Y, Koon AC, Hauser JL, Griffith EC, et al. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron. 2007;53:217–32.
Manabe T, Togashi H, Uchida N, Suzuki SC, Hayakawa Y, Yamamoto M, et al. Loss of cadherin-11 adhesion receptor enhances plastic changes in hippocampal synapses and modifies behavioral responses. Mol Cell Neurosci. 2000;15:534–46.
Evans MM, Kim J, Abel T, Nickl-Jockschat T, Stevens HE. Developmental disruptions of the dorsal striatum in Autism Spectrum Disorder. Biol Psychiatry. 2024;95:102–11.
Walsh JJ, Christoffel DJ, Heifets BD, Ben-Dor GA, Selimbeyoglu A, Hung LW, et al. 5-HT release in nucleus accumbens rescues social deficits in mouse autism model. Nature. 2018;560:589–94.
Bermejo MK, Milenkovic M, Salahpour A, Ramsey AJ. Preparation of synaptic plasma membrane and postsynaptic density proteins using a discontinuous sucrose gradient. J Vis Exp. 2014;3:e51896.
Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45:413–7.
Guo H, Hong S, Jin XL, Chen RS, Avasthi PP, Tu YT, et al. Specificity and efficiency of Cre-mediated recombination in Emx1-Cre knock-in mice. Biochem Biophys Res Commun. 2000;273:661–5.
Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, et al. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–26.
Zerucha T, Stühmer T, Hatch G, Park BK, Long Q, Yu G, et al. A Highly conserved enhancer in the Dlx5/Dlx6 Intergenic Region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20:709–21.
Bauß K, Knapp B, Jores P, Roepman R, Kremer H, Wijk EV, et al. Phosphorylation of the Usher syndrome 1G protein SANS controls Magi2-mediated endocytosis. Hum Mol Genet. 2014;23:3923–42.
Emperador-Melero J, Huson V, van Weering J, Bollmann C, Fischer von Mollard G, Toonen RF, et al. Vti1a/b regulate synaptic vesicle and dense core vesicle secretion via protein sorting at the Golgi. Nat Commun. 2018;9:3421.
Martinez-Garay I. Molecular mechanisms of cadherin function during cortical migration. Front Cell Dev Biol. 2020;8:588152.
Barnes SH, Price SR, Wentzel C, Guthrie SC. Cadherin-7 and cadherin-6B differentially regulate the growth, branching and guidance of cranial motor axons. Development. 2010;137:805–14.
Matcham AC, Toma K, Tsai NY, Sze CJ, Lin PY, Stewart IF, et al. Cadherin-13 maintains retinotectal synapses via transneuronal interactions. J Neurosci. 2024;44:e1310232023.
Schulte JD, Srikanth M, Das S, Zhang J, Lathia JD, Yin L, et al. Cadherin-11 regulates motility in normal cortical neural precursors and glioblastoma. PLoS One. 2013;8:e70962.
Ferri SL, Dow HC, Schoch H, Lee JY, Brodkin ES, Abel T. Age- and sex-specific fear conditioning deficits in mice lacking Pcdh10, an Autism associated gene. Neurobiol Learn Mem. 2021;178:107364.
Nakamoto C, Kawamura M, Nakatsukasa E, Natsume R, Takao K, Watanabe M, et al. GluD1 knockout mice with a pure C57BL/6N background show impaired fear memory, social interaction, and enhanced depressive-like behavior. PLoS One. 2020;15:e0229288.
Mei Y, Monteiro P, Zhou Y, Kim JA, Gao X, Fu Z, et al. Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature. 2016;530:481–4.
Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–7.
Silva-Santos S, van Woerden GM, Bruinsma CF, Mientjes E, Jolfaei MA, Distel B, et al. Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J Clin Invest. 2015;125:2069–76.
Lin S, Fan CY, Wang HR, Li XF, Zeng JL, Lan PX, et al. Frontostriatal circuit dysfunction leads to cognitive inflexibility in neuroligin-3 R451C knockin mice. Mol Psychiatry. 2024;29:2308–20.
Wang C, Derderian KD, Hamada E, Zhou X, Nelson AD, Kyoung H, et al. Impaired cerebellar plasticity hypersensitizes sensory reflexes in SCN2A-associated ASD. Neuron. 2024;112:1444–1455.e5.
Kolling N, Behrens T, Wittmann MK, Rushworth M. Multiple signals in anterior cingulate cortex. Curr Opin Neurobiol. 2016;37:36–43.
Guo B, Chen J, Chen Q, Ren K, Feng D, Mao H, et al. Anterior cingulate cortex dysfunction underlies social deficits in Shank3 mutant mice. Nat Neurosci. 2019;22:1223–34.
Jhang J, Lee H, Kang MS, Lee HS, Park H, Han JH. Anterior cingulate cortex and its input to the basolateral amygdala control innate fear response. Nat Commun. 2018;9:2744.
Gholami-Mahtaj L, Mooziri M, Dehdar K, Abdolsamadi M, Salimi M, Raoufy MR. ACC-BLA functional connectivity disruption in allergic inflammation is associated with anxiety. Sci Rep. 2022;12:3755.
Wang Z, Yueh H, Chau M, Veenstra-VanderWeele J, O’Reilly KC. Circuits underlying social function and dysfunction. Autism Res. 2023;16:1268–88.
Davis GL, Minerva AR, Lario A, Simmler LD, Rodriguez CI, Gunaydin LA. Ketamine increases activity of a fronto-striatal projection that regulates compulsive behavior in SAPAP3 knockout mice. Nat Commun. 2021;12:6040.
Stan A, Pielarski KN, Brigadski T, Wittenmayer N, Fedorchenko O, Gohla A, et al. Essential cooperation of N-cadherin and neuroligin-1 in the transsynaptic control of vesicle accumulation. Proc Natl Acad Sci USA. 2010;107:11116–21.
Dagar S, Teng Z, Gottmann K. Transsynaptic N-cadherin adhesion complexes control presynaptic vesicle and Bulk endocytosis at physiological temperature. Front Cell Neurosci. 2021;15:713693.
Yamagata M, Duan X, Sanes JR. Cadherins interact with synaptic organizers to promote synaptic differentiation. Front Mol Neurosci. 2018;11:142.
Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–69.
Acknowledgements
We thank the ECNU Multifunctional Platform for Innovation (010, 011 and 004) for their mouse breeding service. This work is supported by the National Key Research and Development Program of China 2022YFC2705200, the National Natural Science Foundation of China grant 31100273, 81941013, 32271022, National Natural Science Foundation of China and Israel Science Foundation Cooperative Research Project grant 32061143016 and Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
Y.H. Pan and X. Yuan conceived and supervised the project and obtained the funding. J. Jia, X. Cao, Z. Li, and J. Duan conducted behavioral experiments; X. Cao, J. Jia, J. Duan, and J. Ma conducted histological and molecular analysis; X. Cao conducted the calcium imaging and chemogenetics studies. X. Cao, J. Jia, Z. Li, and Y.H. Pan conducted data analysis and figure organization; X. Cao, Y.H. Pan, and X. Yuan wrote the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of East China Normal University (approval number m20221105) and conducted in accordance with all relevant guidelines and regulations, including the Guide for the Care and Use of Laboratory Animals. This study did not involve human participants.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cao, X., Jia, J., Li, Z. et al. Cadherin 11 regulates presynaptic vesicle trafficking and neuronal activity in autism-related brain circuit. Mol Psychiatry 31, 192–208 (2026). https://doi.org/10.1038/s41380-025-03141-7
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41380-025-03141-7