Abstract
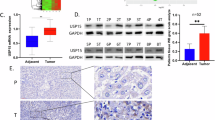
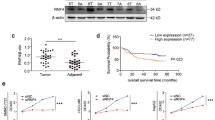
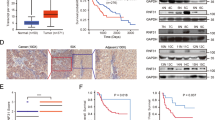
Hepatocellular carcinoma (HCC) is one of the most deadly malignant cancers worldwide. Research into the crucial genes responsible for maintaining the aggressive behaviour of cancer cells is important for the clinical treatment of HCC. The purpose of this study was to determine whether the E3 ubiquitin ligase Ring Finger Protein 125 (RNF125) plays a role in the proliferation and metastasis of HCC. RNF125 expression in human HCC samples and cell lines was investigated using TCGA dataset mining, qRT‒PCR, western blot, and immunohistochemistry assays. In addition, 80 patients with HCC were studied for the clinical value of RNF125. Furthermore, the molecular mechanism by which RNF125 contributes to hepatocellular carcinoma progression was determined with mass spectrometry (MS), coimmunoprecipitation (Co-IP), dual-luciferase reporter assays, and ubiquitin ladder assays. We found that RNF125 was markedly downregulated in HCC tumour tissues, which was associated with a poor prognosis for patients with HCC. Moreover, the overexpression of RNF125 inhibited HCC proliferation and metastasis both in vitro and in vivo, whereas the knockdown of RNF125 exerted antithetical effects. Mechanistically, mass spectrometry analysis revealed a protein interaction between RNF125 and SRSF1, and RNF125 accelerated the proteasome-mediated degradation of SRSF1, which impeded HCC progression by inhibiting the ERK signalling pathway. Furthermore, RNF125 was detected to be the downstream target of miR-103a-3p. In this study, we identified that RNF125 is a tumour suppressor in HCC and inhibits HCC progression by inhibiting the SRSF1/ERK pathway. These findings provide a promising treatment target for HCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6.
Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450–62.
Vogel A, Cervantes A, Chau I, Daniele B, Llovet JM, Meyer T, et al. Hepatocellular carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv238–55.
Roderburg C, Wree A, Demir M, Schmelzle M, Tacke F. The role of the innate immune system in the development and treatment of hepatocellular carcinoma. Hepat Oncol. 2020;7:HEP17.
Ding X, He M, Chan AWH, Song QX, Sze SC, Chen H, et al. Genomic and epigenomic features of primary and recurrent hepatocellular carcinomas. Gastroenterology. 2019;157:1630–45.e6.
Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129–57.
Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18:69–88.
Bijlmakers MJ, Teixeira JM, Boer R, Mayzel M, Puig-Sàrries P, Karlsson G, et al. A C2HC zinc finger is essential for the RING-E2 interaction of the ubiquitin ligase RNF125. Sci Rep. 2016;6:29232.
Yang L, Zhou B, Li X, Lu Z, Li W, Huo X, et al. RNF125 is a ubiquitin-protein ligase that promotes p53 degradation. Cell Physiol Biochem. 2015;35:237–45.
Kim H, Frederick DT, Levesque MP, Cooper ZA, Feng Y, Krepler C, et al. Downregulation of the ubiquitin ligase RNF125 underlies resistance of melanoma cells to BRAF inhibitors via JAK1 deregulation. Cell Rep. 2015;11:1458–73.
Kodama T, Kodama M, Jenkins NA, Copeland NG, Chen HJ, Wei Z. Ring finger protein 125 is an anti-proliferative tumor suppressor in hepatocellular carcinoma. Cancers (Basel). 2022;14:2589.
Vasuri F, Visani M, Acquaviva G, Brand T, Fiorentino M, Pession A, et al. Role of microRNAs in the main molecular pathways of hepatocellular carcinoma. World J Gastroenterol. 2018;24:2647–60.
Komoll RM, Hu Q, Olarewaju O, von Döhlen L, Yuan Q, Xie Y, et al. MicroRNA-342-3p is a potent tumour suppressor in hepatocellular carcinoma. J Hepatol. 2021;74:122–34.
Zhang H, Liu A, Feng X, Tian L, Bo W, Wang H, et al. MiR-132 promotes the proliferation, invasion and migration of human pancreatic carcinoma by inhibition of the tumor suppressor gene PTEN. Prog Biophys Mol Biol. 2019;148:65–72.
Xing S, Tian Z, Zheng W, Yang W, Du N, Gu Y, et al. Hypoxia downregulated miR-4521 suppresses gastric carcinoma progression through regulation of IGF2 and FOXM1. Mol Cancer. 2021;20:9.
Gao JB, Zhu MN, Zhu XL. miRNA-215-5p suppresses the aggressiveness of breast cancer cells by targeting Sox9. FEBS Open Bio. 2019;9:1957–67.
Shimoni-Sebag A, Lebenthal-Loinger I, Zender L, Karni R. RRM1 domain of the splicing oncoprotein SRSF1 is required for MEK1-MAPK-ERK activation and cellular transformation. Carcinogenesis. 2013;34:2498–504.
Das S, Anczuków O, Akerman M, Krainer AR. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 2012;1:110–7.
Wang Y, Xiao X, Zhang J, Choudhury R, Robertson A, Li K, et al. A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat Struct Mol Biol. 2013;20:36–45.
Gonçalves V, Jordan P. Posttranscriptional regulation of splicing factor SRSF1 and its role in cancer cell biology. Biomed Res Int. 2015;2015:287048.
Du JX, Luo YH, Zhang SJ, Wang B, Chen C, Zhu GQ, et al. Splicing factor SRSF1 promotes breast cancer progression via oncogenic splice switching of PTPMT1. J Exp Clin Cancer Res. 2021;40:171.
De Miguel FJ, Sharma RD, Pajares MJ, Montuenga LM, Rubio A, Pio R. Identification of alternative splicing events regulated by the oncogenic factor SRSF1 in lung cancer. Cancer Res. 2014;74:1105–15.
Wu ZH, Liu CC, Zhou YQ, Hu LN, Guo WJ. OnclncRNA-626 promotes malignancy of gastric cancer via inactivated the p53 pathway through interacting with SRSF1. Am J Cancer Res. 2019;9:2249–63.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Yang YM, Kim SY, Seki E. Inflammation and liver cancer: molecular mechanisms and therapeutic targets. Semin Liver Dis. 2019;39:26–42.
Liu Y, Tao S, Liao L, Li Y, Li H, Li Z, et al. TRIM25 promotes the cell survival and growth of hepatocellular carcinoma through targeting Keap1-Nrf2 pathway. Nat Commun. 2020;11:348.
Krais JJ, Wang Y, Bernhardy AJ, Clausen E, Miller JA, Cai KQ, et al. RNF168-mediated ubiquitin signaling inhibits the viability of BRCA1-null cancers. Cancer Res. 2020;80:2848–60.
Chen L, Yuan R, Wen C, Liu T, Feng Q, Deng X, et al. E3 ubiquitin ligase UBR5 promotes pancreatic cancer growth and aerobic glycolysis by downregulating FBP1 via destabilization of C/EBPα. Oncogene. 2021;40:262–76.
Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14.
Yao Q, Chen Y, Zhou X. The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol. 2019;51:11–17.
Khan AQ, Ahmed EI, Elareer NR, Junejo K, Steinhoff M, Uddin S. Role of miRNA-regulated cancer stem cells in the pathogenesis of human malignancies. Cells. 2019;8:840.
Liu Y, Zhang Y, Xiao B, Tang N, Hu J, Liang S, et al. MiR-103a promotes tumour growth and influences glucose metabolism in hepatocellular carcinoma. Cell Death Dis. 2021;12:618.
Sun Z, Zhang Q, Yuan W, Li X, Chen C, Guo Y, et al. MiR-103a-3p promotes tumour glycolysis in colorectal cancer via hippo/YAP1/HIF1A axis. J Exp Clin Cancer Res. 2020;39:250.
Zhang J, Lu Q, Pang H, Zhang M, Wei W. MiR-103a-3p aggravates renal cell carcinoma by targeting TMEM33. Am J Transl Res. 2021;13:12694–703.
Fan Z, Yang J, Zhang D, Zhang X, Ma X, Kang L, et al. The risk variant rs884225 within EGFR impairs miR-103a-3p’s anti-tumourigenic function in non-small cell lung cancer. Oncogene. 2019;38:2291–304.
Wang C, Dong L, Li X, Li Y, Zhang B, Wu H, et al. The PGC1α/NRF1-MPC1 axis suppresses tumor progression and enhances the sensitivity to sorafenib/doxorubicin treatment in hepatocellular carcinoma. Free Radic Biol Med. 2021;163:141–52.
Wang C, Yu H, Lu S, Ke S, Xu Y, Feng Z, et al. LncRNA Hnf4αos exacerbates liver ischemia/reperfusion injury in mice via Hnf4αos/Hnf4α duplex-mediated PGC1α suppression. Redox Biol. 2022;57:102498.
Lu S, Ke S, Wang C, Xu Y, Li Z, Song K, et al. NNMT promotes the progression of intrahepatic cholangiocarcinoma by regulating aerobic glycolysis via the EGFR-STAT3 axis. Oncogenesis. 2022;11:39.
Acknowledgements
The authors would like to thank all the study investigators and staff who participated in this study.
Funding
This work was jointly supported by grants from the Natural Science Foundation of Heilongjiang Province of China (LC2018037), Outstanding Youth Training Fund from Academician Yu Weihan of Harbin Medical University (2014), Scientific Foundation of the First Affiliated Hospital of Harbin Medical University (HYD2020JQ0007, HYD2020JQ0012 and 2019L01), The National Natural Scientific Foundation of China (81100305, 81470876, and 81502605), Heilongjiang Postdoctoral Foundation (LBH-Q17097 and LBH-Z11066), and China Postdoctoral Science Foundation (2012M510990, 2012M520769, and 2013T60387).
Author information
Authors and Affiliations
Contributions
ZF, SK, CW and SL designed and performed experiments, analyzed data and wrote the paper; YX and HY performed experiments and analyzed the data; ZL and BY performed some of the experiments; XL and YH analyzed the data; BQ and MB provided the patient samples for clinical data analysis; YF and YZ provide assistance in the study; YW and YM initiated the study, organized, designed and wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics Approval
The study protocol was approved by the Ethics Committee of The First Affiliated Hospital of Harbin Medical University (2021041; 201823). All animal use and experiments were performed in strict accordance with the procedures approved by the National Cancer Institute Animal Care and Use Committee (ACUC).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Feng, Z., Ke, S., Wang, C. et al. RNF125 attenuates hepatocellular carcinoma progression by downregulating SRSF1-ERK pathway. Oncogene 42, 2017–2030 (2023). https://doi.org/10.1038/s41388-023-02710-w
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-023-02710-w
This article is cited by
-
The enigma of the RING-UIM E3 ligases: its transformative impact on cancer research
Journal of Translational Medicine (2025)
-
miRNA centered regulatory networks identify FN1 and miR27b as metastatic drivers in HPV negative head and neck cancer
Scientific Reports (2025)
-
CircPCNXL2 promotes tumor growth and metastasis by interacting with STRAP to regulate ERK signaling in intrahepatic cholangiocarcinoma
Molecular Cancer (2024)