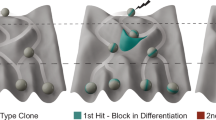
Abstract
Cohesin, a crucial regulator of genome organisation, plays a fundamental role in maintaining chromatin architecture as well as gene expression. Among its subunits, STAG2 stands out because of its frequent deleterious mutations in various cancer types, such as bladder cancer and melanoma. Loss of STAG2 function leads to significant alterations in chromatin structure, disrupts transcriptional regulation, and impairs DNA repair pathways. In this review, we explore the molecular mechanisms underlying cohesin-STAG2 function, highlighting its roles in healthy cells and its contributions to cancer biology, showing how STAG2 dysfunction promotes tumourigenesis and presents opportunities for targeted therapeutic interventions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
16 June 2025
The original online version of this article was revised: Following the publication of this article the authors noted an error in labelling in Figure 1 - specifically the label “SMC1” in the ‘Cohesion Complex’ part of the Figure was duplicated, whereas one of these labels should correctly read “SMC3”.
20 June 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41388-025-03479-w
References
Rouvière-Yaniv J, Yaniv M, Germond J-E. E. coli DNA binding protein HU forms nucleosome-like structure with circular double-stranded DNA. Cell. 1979;17:265–74.
Wolffe AP. Nucleosome positioning and modification: chromatin structures that potentiate transcription. Trends Biochem Sci. 1994;19:240–4.
Dekker J, Heard E. Structural and functional diversity of Topologically Associating Domains. FEBS Lett. 2015;589:2877–84.
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80.
Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5.
Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5.
Rao SSP, Huang S-C, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon K-R, et al. Cohesin loss eliminates all loop domains. Cell. 2017;171:305–20.e324.
Romero-Pérez L, Surdez D, Brunet E, Delattre O, Grünewald TGP. STAG mutations in cancer. Trends Cancer. 2019;5:506–20.
Kojic A, Cuadrado A, De Koninck M, Giménez-Llorente D, Rodríguez-Corsino M, Gómez-López G, et al. Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat Struct Mol Biol. 2018;25:496–504.
Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–114.
Horsfield JA. Full circle: a brief history of cohesin and the regulation of gene expression. FEBS J. 2023;290:1670–87.
Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol. 2000;150:405–16.
Brooker AS, Berkowitz KM. The roles of cohesins in mitosis, meiosis, and human health and disease. Methods Mol Biol. 2014;1170:229–66.
Choi E-H, Yoon S, Koh YE, Hong TK, Do JT, Lee B-K, et al. Meiosis-specific cohesin complexes display essential and distinct roles in mitotic embryonic stem cell chromosomes. Genome Biol. 2022;23:70.
Shen CH, Kim SH, Trousil S, Frederick DT, Piris A, Yuan P, et al. Loss of cohesin complex components STAG2 or STAG3 confers resistance to BRAF inhibition in melanoma. Nat Med. 2016;22:1056–61.
Cuadrado A, Giménez-Llorente D, Kojic A, Rodríguez-Corsino M, Cuartero Y, Martín-Serrano G, et al. Specific contributions of cohesin-SA1 and cohesin-SA2 to TADs and polycomb domains in embryonic stem cells. Cell Rep. 2019;27:3500–10.e3504.
van der Lelij P, Lieb S, Jude J, Wutz G, Santos CP, Falkenberg K, et al. Synthetic lethality between the cohesin subunits STAG1 and STAG2 in diverse cancer contexts. eLife. 2017;6:e26980.
Gligoris T, Löwe J. Structural insights into ring formation of cohesin and related SMC complexes. Trends Cell Biol. 2016;26:680–93.
Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45.
Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57.
Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648.
Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–97.
Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters J-M. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–62.
Gerlich D, Koch B, Dupeux F, Peters J-M, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol. 2006;16:1571–8.
Waizenegger IC, Hauf S, Meinke A, Peters J-M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410.
Weitzer S, Lehane C, Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr Biol. 2003;13:1930–40.
Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14:1598–603.
Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol. 2006;16:863–74.
Hinshaw SM, Makrantoni V, Kerr A, Marston AL, Harrison SC. Structural evidence for Scc4-dependent localization of cohesin loading. eLife. 2015;4:e06057.
Murayama Y, Uhlmann F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature. 2014;505:367–71.
Alonso-Gil D, Cuadrado A, Giménez-Llorente D, Rodríguez-Corsino M, Losada A. Different NIPBL requirements of cohesin-STAG1 and cohesin-STAG2. Nat Commun. 2023;14:1326.
Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, et al. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–37.
Higashi TL, Eickhoff P, Sousa JS, Locke J, Nans A, Flynn HR, et al. A structure-based mechanism for DNA entry into the cohesin ring. Mol Cell. 2020;79:917–33.e919.
Murayama Y, Uhlmann F. DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell. 2015;163:1628–40.
Collier JE, Nasmyth KA. DNA passes through cohesin’s hinge as well as its Smc3–kleisin interface. eLife. 2022;11:e80310.
Minamino M, Ishibashi M, Nakato R, Akiyama K, Tanaka H, Kato Y, et al. Esco1 acetylates cohesin via a mechanism different from that of Esco2. Curr Biol. 2015;25:1694–706.
Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, et al. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143:737–49.
Ladurner R, Kreidl E, Ivanov MP, Ekker H, Idarraga-Amado MH, Busslinger GA, et al. Sororin actively maintains sister chromatid cohesion. EMBO J. 2016;35:635–53.
Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42.
Hellmuth S, Stemmann O. Requirement of Nek2a and cyclin A2 for Wapl-dependent removal of cohesin from prophase chromatin. EMBO J. 2024;0:1–23.
Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–3.
Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–7.
Beckouët F, Hu B, Roig MB, Sutani T, Komata M, Uluocak P, et al. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol Cell. 2010;39:689–99.
Konecna M, Abbasi Sani S, Anger M. Separase and roads to disengage sister chromatids during anaphase. Int J Mol Sci. 2023;24:4604.
Canudas S, Smith S. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol. 2009;187:165–73.
Remeseiro S, Cuadrado A, Carretero M, Martínez P, Drosopoulos WC, Cañamero M, et al. Cohesin‐SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012;31:2076–89.
Kleyman M, Kabeche L, Compton DA. STAG2 promotes error correction in mitosis by regulating kinetochore-microtubule attachments. J Cell Sci. 2014;127:4225–33.
De Koninck M, Lapi E, Badía-Careaga C, Cossío I, Giménez-Llorente D, Rodríguez-Corsino M, et al. Essential roles of cohesin STAG2 in mouse embryonic development and adult tissue homeostasis. Cell Rep. 2020;32:108014.
Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–3.
Mirny LA, Solovei I. Keeping chromatin in the loop(s). Nat Rev Mol Cell Biol. 2021;22:439–40.
Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–95.
Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H. Human cohesin compacts DNA by loop extrusion. Science. 2019;366:1345–9.
Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–33.
Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801.
Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters J-M. DNA loop extrusion by human cohesin. Science. 2019;366:1338–45.
Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–49.
Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 2017;36:3573–99.
Davidson IF, Barth R, Zaczek M, van der Torre J, Tang W, Nagasaka K, et al. CTCF is a DNA-tension-dependent barrier to cohesin-mediated loop extrusion. Nature. 2023;616:822–7.
Nagasaka K, Davidson IF, Stocsits RR, Tang W, Wutz G, Batty P, et al. Cohesin mediates DNA loop extrusion and sister chromatid cohesion by distinct mechanisms. Mol Cell. 2023;83:3049–63.e3046.
Sakata R, Niwa K, Ugarte La Torre D, Gu C, Tahara E, Takada S, et al. Opening of cohesin’s SMC ring is essential for timely DNA replication and DNA loop formation. Cell Rep. 2021;35:108999.
Barth R, Pradhan B, Kim E, Davidson IF, van der Torre J, Peters J-M, et al. Testing pseudotopological and nontopological models for SMC-driven DNA loop extrusion against roadblock-traversal experiments. Sci Rep. 2023;13:8100.
Pradhan B, Barth R, Kim E, Davidson IF, Bauer B, van Laar T, et al. SMC complexes can traverse physical roadblocks bigger than their ring size. Cell Rep. 2022;41:111491.
Davidson IF, Peters JM. Genome folding through loop extrusion by SMC complexes. Nat Rev Mol Cell Biol. 2021;22:445–64.
Li Y, Haarhuis JHI, Sedeño Cacciatore Á, Oldenkamp R, van Ruiten MS, Willems L, et al. The structural basis for cohesin–CTCF-anchored loops. Nature. 2020;578:472–6.
Wutz G, Ladurner R, St Hilaire BG, Stocsits RR, Nagasaka K, Pignard B, et al. ESCO1 and CTCF enable formation of long chromatin loops by protecting cohesin(STAG1) from WAPL. eLife. 2020;9:e52091.
Haarhuis JHI, van der Weide RH, Blomen VA, Yáñez-Cuna JO, Amendola M, van Ruiten MS, et al. The cohesin release factor WAPL restricts chromatin loop extension. Cell. 2017;169:693–707.e614.
Uusküla-Reimand L, Hou H, Samavarchi-Tehrani P, Rudan MV, Liang M, Medina-Rivera A, et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016;17:182.
Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, et al. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–88.
Casa V, Moronta Gines M, Gade Gusmao E, Slotman JA, Zirkel A, Josipovic N, et al. Redundant and specific roles of cohesin STAG subunits in chromatin looping and transcriptional control. Genome Res. 2020;30:515–27.
Cuadrado A, Giménez-Llorente D, De Koninck M, Ruiz-Torres M, Kojic A, Rodríguez-Corsino M, et al. Contribution of variant subunits and associated factors to genome-wide distribution and dynamics of cohesin. Epigenetics Chromatin. 2022;15:37.
Viny AD, Bowman RL, Liu Y, Lavallée VP, Eisman SE, Xiao W, et al. Cohesin members Stag1 and Stag2 display distinct roles in chromatin accessibility and topological control of HSC self-renewal and differentiation. Cell Stem Cell. 2019;25:682–96.e688.
Smith JS, Lappin KM, Craig SG, Liberante FG, Crean CM, McDade SS, et al. Chronic loss of STAG2 leads to altered chromatin structure contributing to de-regulated transcription in AML. J Transl Med. 2020;18:339.
Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, et al. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res. 2012;22:2163–75.
Prekovic S, Schuurman K, Mayayo-Peralta I, Manjón AG, Buijs M, Yavuz S, et al. Glucocorticoid receptor triggers a reversible drug-tolerant dormancy state with acquired therapeutic vulnerabilities in lung cancer. Nat Commun. 2021;12:4360.
Cuartero S, Weiss FD, Dharmalingam G, Guo Y, Ing-Simmons E, Masella S, et al. Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat Immunol. 2018;19:932–41.
Rittenhouse NL, Carico ZM, Liu YF, Stefan HC, Arruda NL, Zhou J, et al. Functional impact of cancer-associated cohesin variants on gene expression and cellular identity. Genetics. 2021;217:iyab025.
Ochi Y, Kon A, Sakata T, Nakagawa MM, Nakazawa N, Kakuta M, et al. Combined cohesin–RUNX1 deficiency synergistically perturbs chromatin looping and causes myelodysplastic syndromes. Cancer Discov. 2020;10:836–53.
Mayayo-Peralta I, Gregoricchio S, Schuurman K, Yavuz S, Zaalberg A, Kojic A, et al. PAXIP1 and STAG2 converge to maintain 3D genome architecture and facilitate promoter/enhancer contacts to enable stress hormone-dependent transcription. Nucleic Acids Res. 2023;51:9576–93.
Wu N, Yu H. The Smc complexes in DNA damage response. Cell Biosci. 2012;2:5.
Caron P, Aymard F, Iacovoni JS, Briois S, Canitrot Y, Bugler B, et al. Cohesin protects genes against γH2AX Induced by DNA double-strand breaks. PLoS Genet. 2012;8:e1002460.
Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–88.
Sjögren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol. 2001;11:991–5.
Gelot C, Guirouilh-Barbat J, Le Guen T, Dardillac E, Chailleux C, Canitrot Y, et al. The cohesin complex prevents the end joining of distant DNA double-strand ends. Mol Cell. 2016;61:15–26.
Arnould C, Rocher V, Finoux A-L, Clouaire T, Li K, Zhou F, et al. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature. 2021;590:660–5.
Yang JH, Brandão HB, Hansen AS. DNA double-strand break end synapsis by DNA loop extrusion. Nat Commun. 2023;14:1913.
Kong X, Ball AR Jr, Pham HX, Zeng W, Chen HY, Schmiesing JA, et al. Distinct functions of human cohesin-SA1 and cohesin-SA2 in double-strand break repair. Mol Cell Biol. 2014;34:685–98.
Zhou J, Nie R-C, He Z-P, Cai X-X, Chen J-W, Lin W-p, et al. STAG2 regulates homologous recombination repair and sensitivity to ATM inhibition. Adv Sci. 2023;10:2302494.
Mondal G, Stevers M, Goode B, Ashworth A, Solomon DA. A requirement for STAG2 in replication fork progression creates a targetable synthetic lethality in cohesin-mutant cancers. Nat Commun. 2019;10:1686.
Countryman P, Fan Y, Gorthi A, Pan H, Strickland E, Kaur P, et al. Cohesin SA2 is a sequence-independent DNA-binding protein that recognizes DNA replication and repair intermediates. J Biol Chem. 2018;293:1054–69.
Bose T, Lee KK, Lu S, Xu B, Harris B, Slaughter B, et al. Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells. PLoS Genet. 2012;8:e1002749.
Harris B, Bose T, Lee KK, Wang F, Lu S, Ross RT, et al. Cohesion promotes nucleolar structure and function. Mol Cell Biol. 2014;25:337–46.
Lawrimore CJ, Bloom K. Common features of the pericentromere and nucleolus. Genes. 2019;10:1029.
Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259.
Meier M, Grant J, Dowdle A, Thomas A, Gerton J, Collas P, et al. Cohesin facilitates zygotic genome activation in zebrafish. Development. 2018;145:dev156521.
Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327:693–6.
Potapova TA, Gerton JL. Ribosomal DNA and the nucleolus in the context of genome organization. Chromosome Res. 2019;27:109–27.
Kim JS, He X, Liu J, Duan Z, Kim T, Gerard J, et al. Systematic proteomics of endogenous human cohesin reveals an interaction with diverse splicing factors and RNA-binding proteins required for mitotic progression. J Biol Chem. 2019;294:8760–72.
Singh AK, Chen Q, Nguyen C, Meerzaman D, Singer DS. Cohesin regulates alternative splicing. Sci Adv. 2023;9:eade3876.
Ule J, Blencowe BJ. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol Cell. 2019;76:329–45.
Porter H, Li Y, Neguembor MV, Beltran M, Varsally W, Martin L, et al. Cohesin-independent STAG proteins interact with RNA and R-loops and promote complex loading. eLife. 2023;12:e79386.
Pan H, Jin M, Ghadiyaram A, Kaur P, Miller HE, Ta HM, et al. Cohesin SA1 and SA2 are RNA binding proteins that localize to RNA containing regions on DNA. Nucleic Acids Res. 2020;48:5639–55.
Alkan F, Wilkins OG, Hernández-Pérez S, Ramalho S, Silva J, Ule J, et al. Identifying ribosome heterogeneity using ribosome profiling. Nucleic Acids Res. 2022;50:e95.
Silva J, Alkan F, Ramalho S, Snieckute G, Prekovic S, Garcia AK, et al. Ribosome impairment regulates intestinal stem cell identity via ZAKɑ activation. Nat Commun. 2022;13:4492.
Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501.
Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45:1459–63.
Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–43.
Balbás-Martínez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Márquez M, Vazquez M, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 2013;45:1464–9.
Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4:1326–41.
Eckardt JN, Stasik S, Röllig C, Sauer T, Scholl S, Hochhaus A, et al. Alterations of cohesin complex genes in acute myeloid leukemia: differential co-mutations, clinical presentation and impact on outcome. Blood Cancer J. 2023;13:18.
Kim JS, He X, Orr B, Wutz G, Hill V, Peters JM, et al. Intact cohesion, anaphase, and chromosome segregation in human cells harboring tumor-derived mutations in STAG2. PLoS Genet. 2016;12:e1005865.
Taylor CF, Platt FM, Hurst CD, Thygesen HH, Knowles MA. Frequent inactivating mutations of STAG2 in bladder cancer are associated with low tumour grade and stage and inversely related to chromosomal copy number changes. Hum Mol Genet. 2014;23:1964–74.
Tothova Z, Valton AL, Gorelov RA, Vallurupalli M, Krill-Burger JM, Holmes A, et al. Cohesin mutations alter DNA damage repair and chromatin structure and create therapeutic vulnerabilities in MDS/AML. JCI Insight. 2021;6:e142149.
Adane B, Alexe G, Seong BKA, Lu D, Hwang EE, Hnisz D, et al. STAG2 loss rewires oncogenic and developmental programs to promote metastasis in Ewing sarcoma. Cancer Cell. 2021;39:827–844.e810.
Chu Z, Gu L, Hu Y, Zhang X, Li M, Chen J, et al. STAG2 regulates interferon signaling in melanoma via enhancer loop reprogramming. Nat Commun. 2022;13:1859.
Richart L, Lapi E, Pancaldi V, Cuenca-Ardura M, Pau EC, Madrid-Mencía M, et al. STAG2 loss-of-function affects short-range genomic contacts and modulates the basal-luminal transcriptional program of bladder cancer cells. Nucleic Acids Res. 2021;49:11005–21.
Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–51.
Boucher A, Murray J, Rao S. Cohesin mutations in acute myeloid leukemia. Leukemia. 2024;38:2318–28.
Montalban-Bravo G, Takahashi K, Alfonso Pierola A, Wang F, Xingzhi S, Jabbour EJ, et al. STAG2 mutations are an independent prognostic factor in patients with myelodysplastic syndromes. Blood. 2016;128:3182.
Thota S, Viny AD, Makishima H, Spitzer B, Radivoyevitch T, Przychodzen B, et al. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood. 2014;124:1790–8.
Bachelot A, Bouvier A, Riou J, Thepot S, Giltat A, Nunes Gomes C, et al. Relationship between comorbidities, mutational profile, and outcome after intensive chemotherapy in patients older than 60 years with acute myeloid leukemia: assessment of different risk scores. Am J Hematol. 2023;98:922–31.
Caballero JC, Dávila J, López-Pavía M, Such E, Bernal T, Ramos F, et al. Outcomes and effect of somatic mutations after erythropoiesis stimulating agents in patients with lower-risk myelodysplastic syndromes. Ther Adv Hematol. 2024;15:20406207231218157.
Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–77.
Lavallée V-P, Gendron P, Boucher G, Lemieux S, Armstrong RN, Boivin I, et al. Mutational and transcriptomic landscape of AML with core-binding factor rearrangements. Blood. 2015;126:802.
Wong WJ, Zon RL, Ho C, Pozdnyakova O, Neuberg DS, Battinelli EM, et al. STAG2 somatic mutations are associated with specific dysplastic megakaryocytic and myeloid cell features in myelodysplastic syndrome. Blood. 2023;142:3233.
Martín-Izquierdo M, Abáigar M, Hernández-Sánchez JM, Tamborero D, López-Cadenas F, Ramos F, et al. Co-occurrence of cohesin complex and Ras signaling mutations during progression from myelodysplastic syndromes to secondary acute myeloid leukemia. Haematologica. 2021;106:2215–23.
Calvo KR, Hickstein DD. The spectrum of GATA2 deficiency syndrome. Blood. 2023;141:1524–32.
West RR, Calvo KR, Embree LJ, Wang W, Tuschong LM, Bauer TR, et al. ASXL1 and STAG2 are common mutations in GATA2 deficiency patients with bone marrow disease and myelodysplastic syndrome. Blood Adv. 2022;6:793–807.
Largeaud L, Collin M, Monselet N, Vergez F, Fregona V, Larcher L, et al. Somatic genetic alterations predict hematological progression in GATA2 deficiency. Haematologica. 2023;108:1515–29.
Fischer A, Hernández-Rodríguez B, Mulet-Lazaro R, Nuetzel M, Hölzl F, van Herk S, et al. STAG2 mutations reshape the cohesin-structured spatial chromatin architecture to drive gene regulation in acute myeloid leukemia. Cell Rep. 2024;43:114498.
Fisher JB, Peterson J, Reimer M, Stelloh C, Pulakanti K, Gerbec ZJ, et al. The cohesin subunit Rad21 is a negative regulator of hematopoietic self-renewal through epigenetic repression of Hoxa7 and Hoxa9. Leukemia. 2017;31:712–9.
Olivieri M, Cho T, Álvarez-Quilón A, Li K, Schellenberg MJ, Zimmermann M, et al. A genetic map of the response to DNA damage in human cells. Cell. 2020;182:481–96.e421.
Wheeler EC, Martin BJE, Doyle WC, Neaher S, Conway CA, Pitton CN, et al. Splicing modulators impair DNA damage response and induce killing of cohesin-mutant MDS and AML. Sci Transl Med. 2024;16:eade2774.
Wan C-L, Liu Y-Q, Liu F-T, Huang Y-H, Cao H-Y, Huang S-M, et al. Venetoclax with hypomethylating agents versus intensive chemotherapy in newly diagnosed acute myeloid leukemia with myelodysplasia related changes: a propensity score-matched analysis based on International Consensus Classification. Blood Cancer J. 2024;14:144.
Weng G, Zhang Y, Yu G, Luo T, Yu S, Xu N, et al. Genetic characteristics predict response to venetoclax plus hypomethylating agents in relapsed or refractory acute myeloid leukemia. J Intern Med. 2023;293:329–39.
Takeda J, Chiba K, Shiraishi Y, Yoshizato T, Shiozawa Y, Nagata Y, et al. Genetic profile of acute erythroid leukemia. Blood. 2016;128:40.
Aquila L, Ohm J, Woloszynska-Read A. The role of STAG2 in bladder cancer. Pharmacol Res. 2018;131:143–9.
Athans S, Krishnan N, Ramakrishnan S, Cortes Gomez E, Lage-Vickers S, Rak M, et al. STAG2 expression is associated with adverse survival outcomes and regulates cell phenotype in muscle-invasive bladder cancer. Cancer Res Commun. 2022;2:1129–43.
Solomon DA, Kim J-S, Bondaruk J, Shariat SF, Wang Z-F, Elkahloun AG, et al. Frequent truncating mutations of STAG2 in bladder cancer. Nat Genet. 2013;45:1428–30.
Taber A, Park Y, Lelo A, Prip F, Xiao J, Berry DL, et al. STAG2 as a prognostic biomarker in low-grade non-muscle invasive bladder cancer. Urol Oncol. 2021;438.e1–438.e9.
Qiao Y, Zhu X, Li A, Yang S, Zhang J. Complete loss of STAG2 expression is an indicator of good prognosis in patients with bladder cancer. Tumor Biol. 2016;37:10279–86.
Gordon NS, Humayun-Zakaria N, Goel A, Abbotts B, Zeegers MP, Cheng KK, et al. STAG2 protein expression in non–muscle-invasive bladder cancer: associations with sex, genomic and transcriptomic changes, and clinical outcomes. Eur Urol Open Sci. 2022;38:88–95.
Lapi E, Kalisz M, Martínez de VJ, Santos C, Sjödahl G, Dyrskjøt L, et al. STAG2 and PPARg as drivers of luminal-type bladder cancer. Urol Oncol. 2020;38:896.
Wang H, Zhong J, Wu C, Liu Y, Zhang J, Zou X, et al. Stromal antigen 2 functions as a tumor suppressor in bladder cancer cells. Oncol Rep. 2017;38:917–25.
Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4:1342–53.
Reed DR, Grohar P, Rubin E, Binitie O, Krailo M, Davis J, et al. Children’s Oncology Group’s 2023 blueprint for research: bone tumors. Pediatr Blood Cancer. 2023;70:e30583.
Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The genomic landscape of the Ewing sarcoma family of tumors reveals recurrent STAG2 mutation. PLOS Genet. 2014;10:e1004475.
Shulman DS, Chen S, Hall D, Nag A, Thorner AR, Lessnick SL, et al. Adverse prognostic impact of the loss of STAG2 protein expression in patients with newly diagnosed localised Ewing sarcoma: a report from the Children’s Oncology Group. Br J Cancer. 2022;127:2220–6.
Surdez D, Zaidi S, Grossetête S, Laud-Duval K, Ferre AS, Mous L, et al. STAG2 mutations alter CTCF-anchored loop extrusion, reduce cis-regulatory interactions and EWSR1-FLI1 activity in Ewing sarcoma. Cancer Cell. 2021;39:810–26.e819.
Sole A, Grossetête S, Heintzé M, Babin L, Zaïdi S, Revy P, et al. Unraveling Ewing sarcoma tumorigenesis originating from patient-derived mesenchymal stem cells. Cancer Res. 2021;81:4994–5006.
El Beaino M, Liu J, Wasylishen AR, Pourebrahim R, Migut A, Bessellieu BJ, et al. Loss of Stag2 cooperates with EWS-FLI1 to transform murine Mesenchymal stem cells. BMC Cancer. 2020;20:3.
Hodis E, Watson Ian R, Kryukov Gregory V, Arold Stefan T, Imielinski M, Theurillat J-P, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63.
Kalaora S, Nagler A, Wargo JA, Samuels Y. Mechanisms of immune activation and regulation: lessons from melanoma. Nat Rev Cancer. 2022;22:195–207.
Oreskovic E, Wheeler EC, Mengwasser KE, Fujimura E, Martin TD, Tothova Z, et al. Genetic analysis of cancer drivers reveals cohesin and CTCF as suppressors of PD-L1. Proc Natl Acad Sci USA. 2022;119:e2120540119.
Juneja VR, McGuire KA, Manguso RT, LaFleur MW, Collins N, Haining WN, et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med. 2017;214:895–904.
Ascierto PA, Kirkwood JM, Grob JJ, Simeone E, Grimaldi AM, Maio M, et al. The role of BRAF V600 mutation in melanoma. J Transl Med. 2012;10:85.
Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71:209–49.
Cai H, Chew SK, Li C, Tsai MK, Andrejka L, Murray CW, et al. A functional taxonomy of tumor suppression in oncogenic KRAS-driven lung cancer. Cancer Discov. 2021;11:1754–73.
Ashkin EL, Tang YJ, Xu H, Hung KL, Belk J, Cai H, et al. A STAG2-PAXIP1/PAGR1 axis suppresses lung tumorigenesis. bioRxiv. 2024. https://doi.org/10.1101/2024.09.14.613043.
Bailey ML, O’Neil NJ, van Pel DM, Solomon DA, Waldman T, Hieter P. Glioblastoma cells containing mutations in the cohesin component STAG2 are sensitive to PARP inhibition. Mol Cancer Ther. 2014;13:724–32.
Xu W, Kim J-S, Yang T, Ya A, Sadzewicz L, Tallon L, et al. STAG2 mutations regulate 3D genome organization, chromatin loops, and Polycomb signaling in glioblastoma multiforme. J Biol Chem. 2024;300:107341.
Li X, Liu Y, Liu J, Qiang W, Ma J, Xie J, et al. STAG2 inactivation reprograms glutamine metabolism of BRAF-mutant thyroid cancer cells. Cell Death Dis. 2023;14:454.
Hong CS, Vasquez JC, Kundishora AJ, Elsamadicy AA, Beckta JM, Sule A, et al. Persistent STAG2 mutation despite multimodal therapy in recurrent pediatric glioblastoma. npj Genom Med. 2020;5:23.
Nie Z, Gao W, Zhang Y, Hou Y, Liu J, Li Z, et al. STAG2 loss-of-function mutation induces PD-L1 expression in U2OS cells. Ann Transl Med. 2019;7:127.
Evers L, Perez-Mancera PA, Lenkiewicz E, Tang N, Aust D, Knösel T, et al. STAG2 is a clinically relevant tumor suppressor in pancreatic ductal adenocarcinoma. Genome Med. 2014;6:9.
Gomes CC, Bernardes VF, Odell EW, Gomez RS. STAG2 loss of expression is rare in aneuploid malignant salivary gland neoplasms. J Oral Pathol Med. 2014;43:273–5.
Bernardes VF, Correa GTB, Loyola AM, Cardoso SV, de Paula AMB, Cabral MMDÁ, et al. STAG2 expression in oral cancer and potentially malignant lesions. Tumor Biol. 2014;35:3641–5.
De Koninck M, Losada A. Cohesin mutations in cancer. Cold Spring Harb Perspect Med. 2016;6:a026476.
Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer. 2018;18:696–705.
Acknowledgements
In Fig. 1, the authors have used the data deposited to Protein Data Base (https://www.rcsb.org) under the designated ID 4pjw (accessed on 08/09/2024). We thank Sarah Vahed for proofreading.
Funding
This work was supported by the KWF/Alpe d’Huzes (14834).
Author information
Authors and Affiliations
Contributions
JSS, LAA, EE, RHS, AM, LJTK, CT and SP contributed to conceptualisation, writing, and editing. SP prepared the figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Scott, J.S., Al Ayadi, L., Epeslidou, E. et al. Emerging roles of cohesin-STAG2 in cancer. Oncogene 44, 277–287 (2025). https://doi.org/10.1038/s41388-024-03221-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-024-03221-y