Abstract
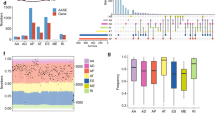
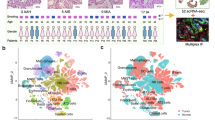
Lung cancer is one of the most frequently diagnosed cancers in the US. African-American (AA) men are more likely to develop lung cancer with higher incidence and mortality rates than European-American (EA) men. Herein, we report high-confidence alternative splicing (AS) events from high-throughput, high-depth total RNA sequencing of lung tumors and non-tumor adjacent tissues (NATs) in two independent cohorts of patients with adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC). We identified novel AS biomarkers with notable differential percent spliced in (PSI) values between lung tumors and NATs enriched in the AA and EA populations, which were associated with oncogenic signaling pathways. We also uncovered tumor subtype- and population-specific AS events associated with cell surface proteins and cancer driver genes. We highlighted significant AS events in SYNE2 specific to LUAD in both populations, as well as those in CD44 from EAs and TMBIM6 from AAs specific to LUAD. Here, we also present the validation of cancer signatures based on direct high-throughput reverse transcription-PCR. Our large survey of lung tumors presents a rich data resource that may help to understand molecular subtypes of lung tumor between AAs and EAs and reveal new therapeutic vulnerabilities that potentially advance health equity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data supporting the findings of this study are available within the article and its Supplementary Information files and from the corresponding authors on request.
Code availability
All scripts for analysis and figure production were built in-house and provided on GitHub (https://github.com/iamsamanzeeshan/LungSplicingAAvsEA).
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA: a cancer J clinicians. 2020;70:7–30.
National Center for Health Statistics (NCHS), Centers for Disease Control and Prevention. 2019; https://www.cdc.gov/nchs/data/nvsr/nvsr70/nvsr70-19.pdf.
Ryan BM. Lung cancer health disparities. Carcinogenesis. 2018;39:741–51.
Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, et al. (eds). SEER Cancer Statistics Review, 1975-2016, National Cancer Institute, Bethesda, MD, https://seer.cancer.gov/csr/1975_2016/, based on November 2018 SEER data submission, posted to the SEER website, April 2019.
Schwartz AG, Wenzlaff AS, Bock CH, Ruterbusch JJ, Chen W, Cote ML, et al. Admixture mapping of lung cancer in 1812 African-Americans. Carcinogenesis. 2011;32:312–7.
Meaney CL, Mitchell KA, Zingone A, Brown D, Bowman E, Yu Y, et al. Circulating inflammation proteins associated with lung cancer in African Americans. J Thorac Oncol: Off Publ Int Assoc Study Lung Cancer. 2019;14:1192–203.
Mitchell KA, Nichols N, Tang W, Walling J, Stevenson H, Pineda M, et al. Recurrent PTPRT/JAK2 mutations in lung adenocarcinoma among African Americans. Nat Commun. 2019;10:5735.
Mitchell KA, Shah E, Bowman ED, Zingone A, Nichols N, Pine SR, et al. Relationship between West African ancestry with lung cancer risk and survival in African Americans. Cancer causes Control: CCC. 2019;30:1259–68.
Meaney CL, Zingone A, Brown D, Yu Y, Cao L, Ryan BM. Identification of serum inflammatory markers as classifiers of lung cancer mortality for stage I adenocarcinoma. Oncotarget. 2017;8:40946–57.
Arauz RF, Byun JS, Tandon M, Sinha S, Kuhn S, Taylor S, et al. Whole-exome profiling of NSCLC among African Americans. J Thorac Oncol. 2020;15:1880–92.
Mitchell KA, Zingone A, Toulabi L, Boeckelman J, Ryan BM. Comparative transcriptome profiling reveals coding and noncoding RNA differences in NSCLC from African Americans and European Americans. Clin Cancer Res. 2017;23:7412–25.
Pine SR, Mechanic LE, Enewold L, Bowman ED, Ryan BM, Cote ML, et al. Differential serum cytokine levels and risk of lung cancer between African and European Americans. Cancer Epidemiol Biomark Prev 2016;25:488–97.
Campbell JD, Lathan C, Sholl L, Ducar M, Vega M, Sunkavalli A, et al. Comparison of prevalence and types of mutations in lung cancers among black and white populations. JAMA Oncol. 2017;3:801–9.
Zingone A, Sinha S, Ante M, Nguyen C, Daujotyte D, Bowman ED, et al. A comprehensive map of alternative polyadenylation in African American and European American lung cancer patients. Nat Commun. 2021;12:5605.
Climente-González H, Porta-Pardo E, Godzik A, Eyras E. The functional impact of alternative splicing in cancer. Cell Rep. 2017;20:2215–26.
Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311–8.
Li Y, Sun N, Lu Z, Sun S, Huang J, Chen Z, et al. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 2017;393:40–51.
Liu Y, Nie H, Liu H, Lu F. Poly(A) inclusive RNA isoform sequencing (PAIso-seq) reveals wide-spread non-adenosine residues within RNA poly(A) tails. Nat Commun. 2019;10:5292.
Kahraman A, Karakulak T, Szklarczyk D, von Mering C. Pathogenic impact of transcript isoform switching in 1209 cancer samples covering 27 cancer types using an isoform-specific interaction network. Sci Rep. 2020;10:14453.
Danan-Gotthold M, Golan-Gerstl R, Eisenberg E, Meir K, Karni R, Levanon EY. Identification of recurrent regulated alternative splicing events across human solid tumors. Nucleic acids Res. 2015;43:5130–44.
Zhang D, Duan Y, Cun J, Yang Q. Identification of prognostic alternative splicing signature in breast carcinoma. Front Genet. 2019;10:278.
Kahles A, Lehmann KV, Toussaint NC, Hüser M, Stark SG, Sachsenberg T, et al. Comprehensive analysis of alternative splicing across tumors from 8705 patients. Cancer cell. 2018;34:211–224.e6.
Bonnal SC, López-Oreja I, Valcárcel J. Roles and mechanisms of alternative splicing in cancer - implications for care. Nat Rev Clin Oncol 2020;17:457–74.
Ding L, Odunsi K. RNA splicing and immune-checkpoint inhibition. N. Engl J Med. 2021;385:1807–9.
Bourcier J, Abdel-Wahab O. Splicing-mediated antigen escape from immunotherapy for B-cell malignancies. Blood Cancer Discov. 2021;3:1–3.
Chang YS, Tu SJ, Chiang HS, Yen JC, Lee YT, Fang HY, et al. Genome-wide analysis of prognostic alternative splicing signature and splicing factors in lung adenocarcinoma. Genes. 2020;11:1300.
Wei C, Xie W, Huang X, Mo X, Liu Z, Wu G, et al. Profiles of alternative splicing events in the diagnosis and prognosis of gastric cancer. J Cancer. 2021;12:2982–92.
Pal S, Bi Y, Macyszyn L, Showe LC, O’Rourke DM, Davuluri RV. Isoform-level gene signature improves prognostic stratification and accurately classifies glioblastoma subtypes. Nucleic acids Res. 2014;42:e64.
Zhu J, Chen Z, Yong L. Systematic profiling of alternative splicing signature reveals prognostic predictor for ovarian cancer. Gynecologic Oncol. 2018;148:368–74.
Zhao W, Hoadley KA, Parker JS, Perou CM. Identification of mRNA isoform switching in breast cancer. BMC genomics. 2016;17:181.
Al Abo M, Hyslop T, Qin X, Owzar K, George DJ, Patierno SR, et al. Differential alternative RNA splicing and transcription events between tumors from African American and White patients in The Cancer Genome Atlas. Genomics. 2021;113:1234–46.
Li S, Hu Z, Zhao Y, Huang S, He X. Transcriptome-wide analysis reveals the landscape of aberrant alternative splicing events in liver cancer. Hepatol. 2019;69:359–75.
Li H, Yang J, Yang G, Ren J, Meng Y, Qi P, et al. Identification of prognostic alternative splicing events in sarcoma. Sci Rep. 2021;11:14949.
Deveaux AE, Allen TA, Al Abo M, Qin X, Zhang D, Patierno BM, et al. RNA splicing and aggregate gene expression differences in lung squamous cell carcinoma between patients of West African and European ancestry. Lung cancer (Amst, Neth). 2021;153:90–98.
Aran D, Camarda R, Odegaard J, Paik H, Oskotsky B, Krings G, et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun. 2017;8:1077.
Sciarrillo R, Wojtuszkiewicz A, Assaraf YG, Jansen G, Kaspers G, Giovannetti E, et al. The role of alternative splicing in cancer: From oncogenesis to drug resistance. Drug resistance updates: Rev commentaries antimicrobial anticancer Chemother. 2020;53:100728.
Robinson TJ, Freedman JA, Al Abo M, Deveaux AE, LaCroix B, Patierno BM, et al. Alternative RNA Splicing as a Potential Major Source of Untapped Molecular Targets in Precision Oncology and Cancer Disparities. Clin Cancer Res: Off J Am Assoc Cancer Res. 2019;25:2963–8.
Fournier G, Cabaud O, Josselin E, Chaix A, Adélaïde J, Isnardon D, et al. Loss of AF6/afadin, a marker of poor outcome in breast cancer, induces cell migration, invasiveness and tumor growth. Oncogene. 2011;30:3862–74.
Smith CC, Selitsky SR, Chai S, Armistead PM, Vincent BG, Serody JS. Alternative tumour-specific antigens. Nat Rev Cancer 2019;19:465–78.
Brosseau JP, Lucier JF, Nwilati H, Thibault P, Garneau D, Gendron D, et al. Tumor microenvironment-associated modifications of alternative splicing. RNA (N. Y, N. Y). 2014;20:189–201.
Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7:e1002218.
Yang Y, Park JW, Bebee TW, Warzecha CC, Guo Y, Shang X, et al. Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the epithelial-to-mesenchymal transition. Mol Cell Biol. 2016;36:1704–19.
Iwasaki J, Kondo T, Darmanin S, Ibata M, Onozawa M, Hashimoto D, et al. FIP1L1 presence in FIP1L1-RARA or FIP1L1-PDGFRA differentially contributes to the pathogenesis of distinct types of leukemia. Ann Hematol. 2014;93:1473–81.
Pintarelli G, Noci S, Maspero D, Pettinicchio A, Dugo M, De Cecco L, et al. Cigarette smoke alters the transcriptome of non-involved lung tissue in lung adenocarcinoma patients. Sci Rep. 2019;9:13039.
Stahre M, Okuyemi KS, Joseph AM, Fu SS. Racial and ethnic differences in the relationship between aspirin use and non-small cell lung cancer risk and survival. Cancer Epidemiol Biomark Prev. 2018;27:1518–26.
Oka M, Xu L, Suzuki T, Yoshikawa T, Sakamoto H, Uemura H, et al. Aberrant splicing isoforms detected by full-length transcriptome sequencing as transcripts of potential neoantigens in non-small cell lung cancer. Genome Biol. 2021;22:9.
Cates HM, Heller EA, Lardner CK, Purushothaman I, Peña CJ, Walker DM, et al. Transcription Factor E2F3a in Nucleus Accumbens Affects Cocaine Action via Transcription and Alternative Splicing. Biol psychiatry. 2018;84:167–79.
Donadoni M, Cicalese S, Sarkar DK, Chang SL, Sariyer IK. Alcohol exposure alters pre-mRNA splicing of antiapoptotic Mcl-1L isoform and induces apoptosis in neural progenitors and immature neurons. Cell death Dis. 2019;10:447.
Lamba V, Yasuda K, Lamba JK, Assem M, Davila J, Strom S, et al. PXR (NR1I2): splice variants in human tissues, including brain, and identification of neurosteroids and nicotine as PXR activators. Toxicol Appl Pharmacol. 2004;199:251–65.
Rajgor D, Mellad JA, Autore F, Zhang Q, Shanahan CM. Multiple novel nesprin-1 and nesprin-2 variants act as versatile tissue-specific intracellular scaffolds. PloS one. 2012;7:e40098.
Hu B, Ma Y, Yang Y, Zhang L, Han H, Chen J. CD44 promotes cell proliferation in non-small cell lung cancer. Oncol Lett. 2018;15:5627–33.
Wang CY, Huang CS, Yang YP, Liu CY, Liu YY, Wu WW, et al. The subpopulation of CD44-positive cells promoted tumorigenicity and metastatic ability in lung adenocarcinoma. J Chin Med Assoc: JCMA. 2019;82:196–201.
Afify AM, Tate S, Durbin-Johnson B, Rocke DM, Konia T. Expression of CD44s and CD44v6 in lung cancer and their correlation with prognostic factors. Int J Biol markers. 2011;26:50–5.
Thapa R, Wilson GD. The importance of CD44 as a stem cell biomarker and therapeutic target in cancer. Stem cells Int. 2016;2016:2087204.
Roberts TC, Langer R, Wood M. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov 2020;19:673–94.
Xiong H, Veedu RN, Diermeier SD. Recent advances in oligonucleotide therapeutics in oncology. Int J Mol Sci. 2021;22:3295.
Anczuków O, Buisson M, Léoné M, Coutanson C, Lasset C, Calender A, et al. BRCA2 deep intronic mutation causing activation of a cryptic exon: opening toward a new preventive therapeutic strategy. Clin Cancer Res: Off J Am Assoc Cancer Res. 2012;18:4903–9.
Peng Q, Zhou Y, Oyang L, Wu N, Tang Y, Su M, et al. Impacts and mechanisms of alternative mRNA splicing in cancer metabolism, immune response, and therapeutics. Molecular therapy: the journal of the American Society of Gene Therapy. 2021; S1525-0016(21)00581-5.
Frankiw L, Baltimore D, Li G. Alternative mRNA splicing in cancer immunotherapy. Nat Rev Immunol 2019;19:675–87.
Lu SX, De Neef E, Thomas JD, Sabio E, Rousseau B, Gigoux M, et al. Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell. 2021;184:4032–4047.e31.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat methods. 2015;12:357–60.
Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111:E5593–E5601.
QIAGEN Ingenuity Pathway Analysis (IPA) https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/.
Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 2018;18:696–705.
Binder JX, Pletscher-Frankild S, Tsafou K, Stolte C, O’Donoghue SI, Schneider R, et al. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database: the journal of biological databases and curation 2014: bau012.
Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinforma (Oxf, Engl). 2014;30:884–6.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids Res. 2015;43:e47.
Funding
This work was supported by a grant from the National Cancer Institute (R01 CA239093) to SRP, HK, BMR, and the Rutgers Cancer Institute of New Jersey Cancer Health Equity Pilot Award to SRP and HK, and Biomedical Informatics Shared Resource at the Rutgers Cancer Institute (P30CA072720-5917).
Author information
Authors and Affiliations
Contributions
BMR conceived and supervised the project, wrote, and revised the manuscript, and acquired funding for this study. SZ designed the study and performed bioinformatic analyzes, including data analysis, writing, and revising the manuscript, and preparing figures. SRP conceived and supervised the project, revised, and edited the manuscript, and acquired funding for this study. HK supervised the project, revised, and edited the manuscript, and acquired the funding for this study. CCH supervised and BD performed the validation experiments. RFA and AZ designed experiments and provided technical and sequencing assistance.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The NCI-MD data that supported the findings of this study were derived from patients enrolled in the NCI-MD case-control study [11]. This study was conducted in accordance with the principles of the Declaration of Helsinki. Institutional review board approval was granted by the National Cancer Institute, Bethesda, Maryland, and the participating hospitals. This study was registered at clinicaltrials.gov [https://clinicaltrials.gov/ct2/show/NCT00339859]. Informed consent was obtained from all participants.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zeeshan, S., Dalal, B., Arauz, R.F. et al. Global profiling of alternative splicing in non-small cell lung cancer reveals novel histological and population differences. Oncogene 44, 958–967 (2025). https://doi.org/10.1038/s41388-024-03267-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-024-03267-y