Abstract
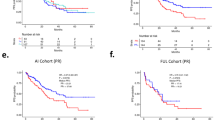
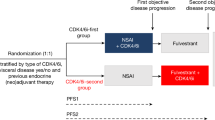
CDK4/6 inhibitors in combination with endocrine therapy are widely used to treat HR+/HER2− metastatic breast cancer leading to improved progression-free survival (PFS) compared to single agent endocrine therapy. Over 300 patients receiving standard-of-care CDK4/6 inhibitor combination therapy for metastatic disease were enrolled at a single institution. Clinical, pathological, and gene expression data were employed to define determinants for PFS duration. Visceral disease (HR 1.55, p = 0.0013), prior endocrine therapy (HR 2.34, p < 0.001), and the type of endocrine therapy (HR 2.16, p < 0.001) were highly associated with PFS duration. Multiple pre-defined gene expression signatures were employed to determine association with response to CDK4/6 inhibitor-based therapy. Random survival forest was applied to define key gene expression and clinical features associated with PFS and develop a predictive model. The time to progression predicted by this model was related to the median PFS observed in PALOMA-2/3 and PEARL studies. Interrogating genes identified as highly significant across all studies indicated common enrichment of gene networks associated with cell cycle and estrogen receptor signaling. These findings indicate that there are common features from real-world use of CDK4/6 inhibitors that could be used to infer time to progression and better inform treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [56] and are accessible through GEO Series accession number GSE285861. Other patient-level data is managed by Roswell Park Comprehensive Cancer Center and is made available through structured data use agreements via their Technology Transfer Office. Investigators may contact the corresponding author to assist with the process of establishing a data use agreement through the Technology Transfer Office.
Code availability
The underlying code for this study is available in the GitHub repository and can be accessed via this link: https://github.com/jianxinwang/ciclib_rsf_predictor.
References
Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA. 2019;321:288–300.
Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G. Breast cancer. Lancet. 2021;397:1750–69.
Gradishar WJ, Moran MS, Abraham J, Abramson V, Aft R, Agnese D, et al. NCCN Guidelines(R) Insights: Breast Cancer, Version 4.2023. J Natl Compr Canc Netw. 2023;21:594–608.
Mouabbi JA, Osborne CK, Schiff R, Rimawi MF. Management of hormone receptor-positive, human epidermal growth factor 2-negative metastatic breast cancer. Breast Cancer Res Treat. 2021;190:189–201.
Carlson RW, Brown E, Burstein HJ, Gradishar WJ, Hudis CA, Loprinzi C, et al. NCCN Task Force Report: Adjuvant Therapy for Breast Cancer. J Natl Compr Canc Netw. 2006;4:S1–26.
Dowsett M, Dunbier AK. Emerging biomarkers and new understanding of traditional markers in personalized therapy for breast cancer. Clin Cancer Res. 2008;14:8019–26.
Sparano JA, Paik S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol. 2008;26:721–8.
Dowsett M, Sestak I, Lopez-Knowles E, Sidhu K, Dunbier AK, Cowens JW, et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol. 2013;31:2783–90.
Cuzick J, Dowsett M, Pineda S, Wale C, Salter J, Quinn E, et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J Clin Oncol. 2011;29:4273–8.
Sotiriou C, Desmedt C. Gene expression profiling in breast cancer. Ann Oncol. 2006;17:x259–62.
Desmedt C, Sotiriou C. Proliferation: the most prominent predictor of clinical outcome in breast cancer. Cell Cycle. 2006;5:2198–202.
Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10:R75.
Ertel A, Dean JL, Rui H, Liu C, Witkiewicz AK, Knudsen KE, et al. RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle. 2010;9:4153–63.
Malorni L, Piazza S, Ciani Y, Guarducci C, Bonechi M, Biagioni C, et al. A gene expression signature of retinoblastoma loss-of-function is a predictive biomarker of resistance to palbociclib in breast cancer cell lines and is prognostic in patients with ER positive early breast cancer. Oncotarget. 2016;7:68012–22.
McCurdy SR, Pacal M, Ahmad M, Bremner R. A CDK2 activity signature predicts outcome in CDK2-low cancers. Oncogene. 2017;36:2491–502.
Sestak I, Buus R, Cuzick J, Dubsky P, Kronenwett R, Denkert C, et al. Comparison of the Performance of 6 Prognostic Signatures for Estrogen Receptor-Positive Breast Cancer: A Secondary Analysis of a Randomized Clinical Trial. JAMA Oncol. 2018;4:545–53.
Sinn P, Aulmann S, Wirtz R, Schott S, Marme F, Varga Z, et al. Multigene Assays for Classification, Prognosis, and Prediction in Breast Cancer: a Critical Review on the Background and Clinical Utility. Geburtshilfe Frauenheilkd. 2013;73:932–40.
Prat A, Parker JS, Fan C, Cheang MC, Miller LD, Bergh J, et al. Concordance among gene expression-based predictors for ER-positive breast cancer treated with adjuvant tamoxifen. Ann Oncol. 2012;23:2866–73.
Albain KS, Paik S, van’t Veer L. Prediction of adjuvant chemotherapy benefit in endocrine responsive, early breast cancer using multigene assays. Breast. 2009;18:S141–5.
Paik S. Development and clinical utility of a 21-gene recurrence score prognostic assay in patients with early breast cancer treated with tamoxifen. Oncologist. 2007;12:631–5.
Robertson JFR, Bondarenko IM, Trishkina E, Dvorkin M, Panasci L, Manikhas A, et al. Fulvestrant 500 mg versus anastrozole 1 mg for hormone receptor-positive advanced breast cancer (FALCON): an international, randomised, double-blind, phase 3 trial. Lancet. 2016;388:2997–3005.
McAndrew NP, Finn RS. Clinical Review on the Management of Hormone Receptor-Positive Metastatic Breast Cancer. JCO Oncol Pr. 2022;18:319–27.
Nagaraj G, Ma CX. Clinical Challenges in the Management of Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Metastatic Breast Cancer: A Literature Review. Adv Ther. 2021;38:109–36.
Gao JJ, Cheng J, Bloomquist E, Sanchez J, Wedam SB, Singh H, et al. CDK4/6 inhibitor treatment for patients with hormone receptor-positive, HER2-negative, advanced or metastatic breast cancer: a US Food and Drug Administration pooled analysis. Lancet Oncol. 2020;21:250–60.
Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16:25–35.
Cristofanilli M, Turner NC, Bondarenko I, Ro J, Im SA, Masuda N, et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol. 2016;17:425–39.
Alvarez-Fernandez M, Malumbres M. Mechanisms of Sensitivity and Resistance to CDK4/6 Inhibition. Cancer Cell. 2020;37:514–29.
Knudsen ES, Witkiewicz AK. The Strange Case of CDK4/6 Inhibitors: Mechanisms, Resistance, and Combination Strategies. Trends Cancer. 2017;3:39–55.
Freeman-Cook K, Hoffman RL, Miller N, Almaden J, Chionis J, Zhang Q, et al. Expanding control of the tumor cell cycle with a CDK2/4/6 inhibitor. Cancer Cell. 2021;39:1404–21 e11.
Prat A, Chaudhury A, Solovieff N, Pare L, Martinez D, Chic N, et al. Correlative Biomarker Analysis of Intrinsic Subtypes and Efficacy Across the MONALEESA Phase III Studies. J Clin Oncol. 2021;39:1458–67.
Guerrero-Zotano A, Belli S, Zielinski C, Gil-Gil M, Fernandez-Serra A, Ruiz-Borrego M, et al. CCNE1 and PLK1 mediates resistance to palbociclib in HR+/HER2- metastatic breast cancer. Clin Cancer Res. 2023;29:1557–68.
Zhu Z, Turner NC, Loi S, André F, Martin M, Diéras V, et al. Comparative biomarker analysis of PALOMA-2/3 trials for palbociclib. NPJ Precis Oncol. 2022;6:56.
Park YH, Im SA, Park K, Wen J, Lee KH, Choi YL, et al. Longitudinal multi-omics study of palbociclib resistance in HR-positive/HER2-negative metastatic breast cancer. Genome Med. 2023;15:55.
Witkiewicz AK, Schultz E, Wang J, Hamilton D, Levine E, O’Connor T, et al. Determinants of response to CDK4/6 inhibitors in the real-world setting. NPJ Precis Oncol. 2023;7:90.
Knudsen ES, Schultz E, Hamilton D, Attwood K, Edge S, O’Connor T, et al. Real-World Experience with CDK4/6 Inhibitors for Metastatic HR+/HER2- Breast Cancer at a Single Cancer Center. Oncologist. 2022;27:646–54.
Chia SK, Bramwell VH, Tu D, Shepherd LE, Jiang S, Vickery T, et al. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin Cancer Res. 2012;18:4465–72.
Ishwaran H, Gerds TA, Kogalur UB, Moore RD, Gange SJ, Lau BM. Random survival forests for competing risks. Biostatistics. 2014;15:757–73.
Chen X, Ishwaran H. Pathway hunting by random survival forests. Bioinformatics. 2013;29:99–105.
Mogensen UB, Ishwaran H, Gerds TA. Evaluating Random Forests for Survival Analysis using Prediction Error Curves. J Stat Softw. 2012;50:1–23.
Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–34.
Rosario SR, Long MD, Affronti HC, Rowsam AM, Eng KH, Smiraglia DJ. Pan-cancer analysis of transcriptional metabolic dysregulation using The Cancer Genome Atlas. Nat Commun. 2018;9:5330.
Knudsen ES, Nambiar R, Rosario SR, Smiraglia DJ, Goodrich DW, Witkiewicz AK. Pan-cancer molecular analysis of the RB tumor suppressor pathway. Commun Biol. 2020;3:158.
Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77.
Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 2010;29:4018–32.
Condorelli R, Spring L, O’Shaughnessy J, Lacroix L, Bailleux C, Scott V, et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol. 2018;29:640–5.
O’Leary B, Cutts RJ, Liu Y, Hrebien S, Huang X, Fenwick K, et al. The Genetic Landscape and Clonal Evolution of Breast Cancer Resistance to Palbociclib plus Fulvestrant in the PALOMA-3 Trial. Cancer Discov. 2018;8:1390–403.
O’Brien RC, Ishwaran H, Szczotka-Flynn LB, Lass JH. Cornea Preservation Time Study G. Random Survival Forests Analysis of Intraoperative Complications as Predictors of Descemet Stripping Automated Endothelial Keratoplasty Graft Failure in the Cornea Preservation Time Study. JAMA Ophthalmol. 2021;139:191–7.
O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13:417–30.
Ma CX, Gao F, Luo J, Northfelt DW, Goetz MP, Forero A, et al. NeoPalAna: Neoadjuvant palbociclib, a cyclin-dependent kinase 4/6 inhibitor, and anastrozole for clinical stage 2 or 3 estrogen receptor positive breast cancer. Clin Cancer Res. 2017;23:4055–65.
Mosley JD, Keri RA. Cell cycle correlated genes dictate the prognostic power of breast cancer gene lists. BMC Med Genomics. 2008;1:11.
Turner NC, Liu Y, Zhu Z, Loi S, Colleoni M, Loibl S, et al. Cyclin E1 Expression and Palbociclib Efficacy in Previously Treated Hormone Receptor-Positive Metastatic Breast Cancer. J Clin Oncol. 2019;37:1169–78.
Yang C, Li Z, Bhatt T, Dickler M, Giri D, Scaltriti M, et al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene. 2017;36:2255–64.
Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene Expression and Benefit of Chemotherapy in Women With Node-Negative, Estrogen Receptor-Positive Breast Cancer. J Clin Oncol. 2023;41:3565–75.
Guerrero-Zotano Á, Belli S, Zielinski C, Gil-Gil M, Fernandez-Serra A, Ruiz-Borrego M, et al. CCNE1 and PLK1 Mediate Resistance to Palbociclib in HR+/HER2- Metastatic Breast Cancer. Clin Cancer Res. 2023;29:1557–68.
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inf. 2009;42:377–81.
Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10.
Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9.
Zhang Y, Schnabel CA, Schroeder BE, Jerevall PL, Jankowitz RC, Fornander T, et al. Breast cancer index identifies early-stage estrogen receptor-positive breast cancer patients at risk for early- and late-distant recurrence. Clin Cancer Res. 2013;19:4196–205.
Filipits M, Rudas M, Jakesz R, Dubsky P, Fitzal F, Singer CF, et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin Cancer Res. 2011;17:6012–20.
van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009.
Bartlett JM, Thomas J, Ross DT, Seitz RS, Ring BZ, Beck RA, et al. Mammostrat as a tool to stratify breast cancer patients at risk of recurrence during endocrine therapy. Breast Cancer Res. 2010;12:R47.
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26.
Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Acknowledgements
The authors would like to thank the patients for their participation in this study. We acknowledge the entire breast oncology group at Roswell Park for their assistance in carrying out this study and Ms. Deanna Hamilton for her assistance in coordinating and collecting data for the protocol. This study was supported by funding from the Roswell Park Alliance Foundation and grants to ESK and AKW from the NCI (CA247362 and CA247362-S1).
Author information
Authors and Affiliations
Contributions
The study was designed by AKW and ESK. Data was collected by ES, TO, EL, and AKW. Coordination of tissue collection and analysis for this study was performed by AKW and ES. Data was analyzed by JW, AKW, ESK, ES, and TNO. The manuscript was assembled and written by ESK, TNO, ES, JW, and AKW. All authors read, edited, and approved the final manuscript. Funding for this study was attained by AKW and ESK.
Corresponding authors
Ethics declarations
Competing interests
ESK and AKW have sponsored research funded by Blueprint Medicine and Bristol Myers Squibb, in addition to funding received from the Roswell Park Alliance Foundation and National Cancer Institute. ESK is also a member of the Cancer Cell Cycle-LLC consulting enterprise. All other authors have no competing interests to declare.
Ethics approval and consent to participate
All patient data collection, protocols, and methods of this specific study were conducted in accordance with the principles outlined in the Declaration of Helsinki. All methods used in this study were approved by the RPCCC Institutional Review Board under the Roswell Park Remnant Tissue Protocol and through written informed consent received from all participating patients under clinical trial NCT04526587.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Witkiewicz, A.K., Wang, J., Schultz, E. et al. Using prognostic signatures and machine learning to identify core features associated with response to CDK4/6 inhibitor-based therapy in metastatic breast cancer. Oncogene 44, 1387–1399 (2025). https://doi.org/10.1038/s41388-025-03308-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03308-0