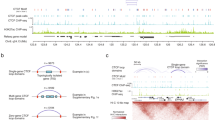
Abstract
Alternative splicing is a fundamental mechanism that generates functionally distinct proteins from individual genes, contributing to gene regulation and proteomic diversity. In Drosophila, the bunched (bun) gene, a member of the TSC-22 domain gene family, undergoes alternative splicing, yielding diverse protein isoforms involved in crucial biological processes. Nevertheless, the specific roles and regulatory mechanisms of each isoform remain elusive. Here, we employed CRISPR/Cas9 technology to introduce targeted deletions within the endogenous locus of the bun gene, resulting in the removal of either long or short isoforms. We discovered that the short isoforms demonstrated a growth-suppressive role, whereas the long isoforms exhibited a growth-promoting effect. Surprisingly, the long isoforms exhibited a remarkable dual functionality, as both deletion and amplification of long isoform expression impede the excess growth induced by Hippo pathway inactivation. Mechanistically, ectopically expressed Bun long isoforms act as the transcriptional suppressor by competitively binding to targets’ promoter regions in conjunction with Yorkie/Scalloped (Yki/Sd), thereby inhibiting its transcriptional outputs and ultimately leading to the growth suppression. These findings unveil the intricate interaction between distinct spliced isoforms of Bun and oncogenic outcomes, highlighting Bun long isoforms as the critical transcription suppressor regulating Hippo pathway inactivation-mediated growth and tumorigenesis in Drosophila.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Raw sequencing data for the three BunA CUT&Tag replicates has been submitted to the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE274246) under the accession number of GSE274246.
References
Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–7.
Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–63.
Sammeth M, Foissac S, Guigo R. A general definition and nomenclature for alternative splicing events. PLoS Comput Biol. 2008;4:e1000147.
Srivastava D, de Toledo M, Manchon L, Tazi J, Juge F. Modulation of Yorkie activity by alternative splicing is required for developmental stability. EMBO J. 2021;40:e104895.
Tsang M-J, Cheeseman IM. Alternative CDC20 translational isoforms tune mitotic arrest duration. Nature. 2023;617:154–61.
Su Z, Huang D. Alternative splicing of Pre-mRNA in the control of immune activity. Genes. 2021;12:574.
Jones L, Naidoo M, Machado LR, Anthony K. The Duchenne muscular dystrophy gene and cancer. Cell Oncol. 2020;44:19–32.
Ladomery MR, Harper SJ, Bates DO. Alternative splicing in angiogenesis: the vascular endothelial growth factor paradigm. CANCER LETTERS. 2007;249:133–42.
Shao Y, Chong W, Liu X, Xu Y, Zhang H, Xu Q, et al. Alternative splicing-derived intersectin1-L and intersectin1-S exert opposite function in glioma progression. Cell Death Dis. 2019;10:431.
Boise LH, GonzBlez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, et al. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608.
Boeckx B, Shahi RB, Smeets D, De Brakeleer S, Decoster L, Van Brussel T, et al. The genomic landscape of nonsmall cell lung carcinoma in never smokers. Int J Cancer. 2020;146:3207–18.
Xu XQ, Sun R, Li YZ, Wang JX, Zhang M, Xiong X, et al. Comprehensive bioinformatic analysis of the expression and prognostic significance of TSC22D domain family genes in adult acute myeloid leukemia. BMC Med Genomics. 2023;16:117.
Nakamura M, Kitaura J, Enomoto Y, Lu Y, Nishimura K, Isobe M, et al. Transforming growth factor-β-stimulated clone-22 is a negative-feedback regulator of Ras/Raf signaling: Implications for tumorigenesis. Cancer Sci. 2012;103:26–33.
Huser CA, Pringle MA, Heath VJ, Bell AK, Kendrick H, Smalley MJ, et al. TSC-22D1 isoforms have opposing roles in mammary epithelial cell survival. Cell Death Differ. 2010;17:304–15.
Gluderer S, Oldham S, Rintelen F, Sulzer A, Schütt C, Wu X, et al. Bunched, the Drosophila homolog of the mammalian tumor suppressor TSC-22, promotes cellular growth. BMC developmental Biol. 2008;8:10.
Shibanuma M, Kuroki T, Nose K. Isolation of a gene encoding a putative leucine zipper structure that is induced by transforming growth factor beta 1 and other growth factors. J Biol Chem. 1992;267:10219–24.
Kester HA, Blanchetot C, den Hertog J, van der Saag PT, van der Burg B. Transforming growth factor-beta-stimulated clone-22 is a member of a family of leucine zipper proteins that can homo- and heterodimerize and has transcriptional repressor activity. J Biol Chem. 1999;274:27439–47.
Landschulz WH, Johnson PF, McKnight SL. The Leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–64.
O’Shea EK, Rutkowski R, Kim PS. Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell. 1992;68:699–708.
Treisman JE, Lai Z-C, Rubin GM. shortsighted acts in the decapentaplegic pathway in Drosophila eye development and has homology to a mouse TGF-β-responsive gene. Development. 1995;121:2835–45.
Dobens LL. Drosophila bunched integrates opposing DPP and EGF signals to set the operculum boundary. Dev Genes Evolut. 2000;127:745–54.
Wu X, Yamada-Mabuchi M, Morris EJ, Tanwar PS, Dobens L, Gluderer S, et al. The Drosophila homolog of human tumor suppressor TSC-22 promotes cellular growth, proliferation, and survival. Proc Natl Acad Sci USA. 2008;105:5414–9.
Gluderer S, Brunner E, Germann M, Jovaisaite V, Li C, Rentsch CA, et al. Madm (Mlf1 adapter molecule) cooperates with Bunched A to promote growth in Drosophila. J Biol. 2010;9:9.
Ren X, Yang Z, Xu J, Sun J, Mao D, Hu Y, et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014;9:1151–62.
Oh H, Irvine KD. in vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–8.
Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science. 2003;14:1227–31.
Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8.
Brumby AM, Richardson HE. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J. 2003;3:5769–79.
Humbert PO, Grzeschik NA, Brumby AM, Galea R, Elsum I, Richardson HE. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 2008;27:6888–907.
Ma X, Lu JY, Dong Y, Li D, Malagon JN, Xu T. PP6 disruption synergizes with oncogenic Ras to promote JNK-dependent tumor growth and invasion. Cell Rep. 2017;19:2657–64.
Ma X, Lu JY, Moraru A, Teleman AA, Fang J, Qiu Y, et al. A novel regulator of ER Ca(2+) drives Hippo-mediated tumorigenesis. Oncogene. 2020;39:1378–87.
Fish MP, Groth AC, Calos MP, Nusse R. Creating transgenic Drosophila by microinjecting the site-specific φC31 integrase mRNA and a transgene-containing donor plasmid. Nat Protoc. 2007;2:2325–31.
Ohta S, Shimekake Y, Nagata K. Molecular cloning and characterization of a transcription factor for the C-type natriuretic peptide gene promoter. Eur J Biochem. 1996;242:460–6.
Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gústafsdóttir SM, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–7.
Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor scalloped mediates hippo signaling in organ size control. Dev Cell. 2008;14:377–87.
Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–34.
Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, et al. The FERM-domain protein expanded regulates hippo pathway activity via direct interactions with the transcriptional activator yorkie. Dev Cell. 2009;16:411–20.
Zhang X, Milton CC, Poon CLC, Hong W, Harvey KF. Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador-Warts-Hippo pathway. Cell Death Differ. 2011;18:1346–55.
Nie Y, Li Q, Amcheslavsky A, Duhart JC, Veraksa A, Stocker H, et al. Bunched and madm function downstream of tuberous sclerosis complex to regulate the growth of intestinal stem cells in drosophila. Stem cell Rev Rep. 2015;11:813–25.
Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–20.
Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, et al. The tumor-suppressor genes NF2/Merlin and expanded act through Hippo signaling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36.
Chen C-L, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci. 2010;107:15810–5.
El Marabti E, Younis I. The cancer spliceome: reprograming of alternative splicing in cancer. Front Mol Biosci. 2018;5:80.
Hattori D, Demir E, Kim HW, Viragh E, Zipursky SL, Dickson BJ. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. 2007;449:223–7.
Marasco LE, Kornblihtt AR. The physiology of alternative splicing. Nat Rev Mol Cell Biol. 2022;24:242–54.
Seidel G, Adermann K, Schindler T, Ejchart A, Jaenicke R, Forssmann W-G, et al. Solution structure of porcine delta sleep-inducing peptide immunoreactive peptide a homolog of the shortsightedgene product. J Biol Chem. 1997;272:30918–27.
Campbell S, Inamdar M, Rodrigue V, Raghavan V, Palazzolo M, Chovnick A. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes Dev. 1992;6:367–79.
Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber JL, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–41.
Pietenpol JA, Holt JT, Stein RW, Moses HL. Transforming growth factor beta 1 suppression of c-myc gene transcription: role in inhibition of keratinocyte proliferation. Proc Natl Acad Sci USA. 1990;87:3758–62.
Acknowledgements
We thank Tian Xu, Lei Xue, Bloomington Drosophila Stock Center, Vienna Drosophila Resource Center, TsingHua Fly Center, and DSHB for providing fly stocks and reagents. We thank Wenhan Liu from our lab for fly stock maintenance. Cartoon illustrations in the manuscript were created with BioRender.com. This work was supported by startup funds from Westlake University9uj and grants from the National Natural Science Foundation of China (32170824, 32322027) to XM and (32370710) to YZ, HRHI program (1011103360222B1) of Westlake Laboratory of Life Sciences and Biomedicine to XM, Westlake Laboratory of Life Sciences and Biomedicine (10128A092001), and “Team for Growth Control and Size Innovative Research” (201804016). YZ is also supported by the National Key Research and Development Program (2022YFA1302700).
Author information
Authors and Affiliations
Contributions
XM conceived and designed the study; XM, YZ, and PG designed the experiments and analyzed the data; PG performed the majority of the experiments with the help and input from SS, DZ, XK, and ZZ; YN performed CUT&Tag analyses; XM, SS, XK, and PG wrote the original manuscript; XM, PG, and SS wrote the revised manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The authors confirm that all methods were performed in accordance with the relevant guidelines and regulations. All animal procedures were performed following the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Westlake University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guo, P., Song, S., Niu, Y. et al. Alternative splicing of bunched confers a dual role in hippo pathway-dependent growth and tumorigenesis. Oncogene 44, 1949–1960 (2025). https://doi.org/10.1038/s41388-025-03348-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03348-6