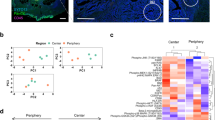
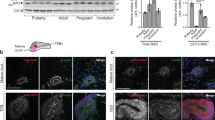
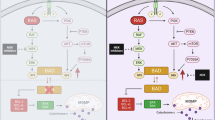
Abstract
The Bcl-2 family member BAD is a candidate disease modulator because it stimulates apoptosis in a cell context basis and inhibits cell migration during normal mammary gland morphogenesis. This activity depends on 3 key regulatory serines (S75, 99, 118) in the unphosphorylated state. Given that developmental programs are often hijacked in cancer, we hypothesized that BAD would impede breast cancer progression. We generated breast cancer mouse models representing loss-of-function or phosphorylation deficient mutations (PyMT-Bad−/− and PyMT-Bad3SA/3SA, respectively). Preventing BAD phosphorylation significantly decreased breast cancer progression and metastasis. The knock-out phenocopied the control PyMT-Bad+/+ suggesting that phosphorylated BAD protein was inert. Thus, the BAD3SA mutation unmasked latent anti-tumor activity. Indeed, transcriptomics showed PyMT-Bad3SA/3SA activated multiple anti-tumor programs including apoptosis, inflammation, cellular differentiation, and diminished cell migration. This anti-tumor effect associated with clinical survival of breast cancer patients whose tumors had high levels of unphosphorylated BAD. Kinase screens identified ERK as the major BAD kinase in breast cells, and ERK inhibition impeded tumoroid invasion. Our data suggest that unphosphorylated BAD modulates anti-tumor pathways that contribute to excellent patient prognosis. Thus, targeting ERK to dephosphorylate BAD may be an exciting therapeutic opportunity in the future.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Uncropped raw blots underlying the figures are in Supplementary Table 4. RNA-seq raw paired-end sequencing reads (fastq format) and complete DESeq2 analysis are deposited in the Gene Expression Omnibus (GEO) public repository, accession GSE196626. Main functions, packages, and modules used in MATLAB(version R2022a/b), R(version 4.2.2), and Python (version 3.9,) respectively, as well as bi-directional communication and data exchange between MATLAB and R are detailed in the ‘Methods’ and Supplementary File 1. Detailed custom codes and their dependency to reproduce the data are included in Supplementary File 1.
References
Meirson T, Gil-Henn H, Samson AO. Invasion and metastasis: the elusive hallmark of cancer. Oncogene. 2020;39:2024–6.
Giaquinto AN, Sung H, Miller KD, Kramer JL, Newman LA, Minihan A, et al. Breast Cancer Statistics, 2022. CA Cancer J Clin. 2022;72:524–41.
Pierce GB, Speers WC. Tumors as caricatures of the process of tissue renewal: prospects for therapy by directing differentiation. Cancer Res. 1988;48:1996–2004.
Condeelis J, Singer RH, Segall JE. The great escape: when cancer cells hijack the genes for chemotaxis and motility. Annu Rev Cell Dev Biol. 2005;21:695–718.
Githaka JM, Pirayeshfard L, Goping IS. Cancer invasion and metastasis: Insights from murine pubertal mammary gland morphogenesis. Biochim Biophys Acta Gen Subj 2023: 130375.
Githaka JM, Tripathi N, Kirschenman R, Patel N, Pandya V, Kramer DA, et al. BAD regulates mammary gland morphogenesis by 4E-BP1-mediated control of localized translation in mouse and human models. Nat Commun. 2021;12:2939.
Danial NN. BAD: Undertaker by night, candyman by day. Oncogene 2008; 27: 53.
Boac BM, Abbasi F, Ismail-Khan R, Xiong Y, Siddique A, Park H, et al. Expression of the BAD pathway is a marker of triple-negative status and poor outcome. Sci Rep. 2019;9:17496.
Craik AC, Veldhoen RA, Czernick M, Buckland TW, Kyselytzia K, Ghosh S, et al. The BH3-only protein Bad confers breast cancer taxane sensitivity through a nonapoptotic mechanism. Oncogene. 2010;29:5381–91.
Mann J, Githaka JM, Buckland TW, Yang N, Montpetit R, Patel N, et al. Non-canonical BAD activity regulates breast cancer cell and tumor growth via 14-3-3 binding and mitochondrial metabolism. Oncogene. 2019;38:3325–39.
Mann J, Yang N, Montpetit R, Kirschenman R, Lemieux H, Goping IS. BAD sensitizes breast cancer cells to docetaxel with increased mitotic arrest and necroptosis. Sci Rep. 2020;10:355.
Datta SR, Ranger AM, Lin MZ, Sturgill JF, Ma Y-C, Cowan CW, et al. Survival factor-mediated BAD phosphorylation raises the mitochondrial threshold for apoptosis. Dev Cell. 2002;3:631–43.
Padmanaban V, Grasset EM, Neumann NM, Fraser AK, Henriet E, Matsui W, et al. Organotypic culture assays for murine and human primary and metastatic-site tumors. Nat Protoc 2020;15. https://doi.org/10.1038/s41596-020-0335-3.
Lerner EP, Arjomand Fard N, Githaka JM, Hotte N, Ezeh C, Huynh HQ, et al. Establishment of a National Surgical Tissue Biobank for Pediatric Crohn’s Disease: An Implementation Feasibility Study. J Pediatr Surg 2025;60. https://doi.org/10.1016/J.JPEDSURG.2025.162195.
Pandya V, Githaka JM, Patel N, Veldhoen R, Hugh J, Damaraju S, et al. BIK drives an aggressive breast cancer phenotype through sublethal apoptosis and predicts poor prognosis of ER-positive breast cancer. Cell Death Dis. 2020;11:1–19.
Githaka JM, Vega AR, Baird MA, Davidson MW, Jaqaman K, Touret N. Ligand-induced growth and compaction of CD36 nanoclusters enriched in Fyn induces Fyn signaling. J Cell Sci. 2016;129:4175–89.
Githaka JM. Molnupiravir does not induce mutagenesis in host lung cells during SARS-CoV-2 treatment. Bioinform Biol Insights. 2022;16:11779322221085076.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15. https://doi.org/10.1186/S13059-014-0550-8.
Zare A, Postovit L-M, Githaka JM. Robust inflammatory breast cancer gene signature using nonparametric random forest analysis. Breast Cancer Res. 2021;23:1–6.
Gendoo DMA, Ratanasirigulchai N, Schröder MS, Paré L, Parker JS, Prat A, et al. Genefu: an R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics. 2016;32:1097–9.
Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35:i436–i445.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:90.
Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–61.
Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–26.
Ursini-Siegel J, Hardy WR, Zuo D, Lam SHL, Sanguin-Gendreau V, Cardiff RD, et al. ShcA signalling is essential for tumour progression in mouse models of human breast cancer. EMBO J. 2008;27:910–20.
Hong G, Zhang W, Li H, Shen X, Guo Z. Separate enrichment analysis of pathways for up- and downregulated genes. J R Soc Interface 2013;11. https://doi.org/10.1098/RSIF.2013.0950.
Cassier PA, Navaridas R, Bellina M, Rama N, Ducarouge B, Hernandez-Vargas H, et al. Netrin-1 blockade inhibits tumour growth and EMT features in endometrial cancer. Nature. 2023;620:409–16.
Fitamant J, Guenebeaud C, Coissieux MM, Guix C, Treilleux I, Scoazec JY, et al. Netrin-1 expression confers a selective advantage for tumor cell survival in metastatic breast cancer. Proc Natl Acad Sci USA. 2008;105:4850–5.
Ewald AJ, Huebner RJ, Palsdottir H, Lee JK, Perez MJ, Jorgens DM, et al. Mammary collective cell migration involves transient loss of epithelial features and individual cell migration within the epithelium. J Cell Sci. 2012;125:2638–54.
Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–88.
Gorringe KL, Fox SB. Ductal carcinoma in situ biology, biomarkers, and diagnosis. Front Oncol 2017; 7. https://doi.org/10.3389/FONC.2017.00248.
Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14:570–81.
Wang Z, Sandiford S, Wu C, Li SSC. Numb regulates cell-cell adhesion and polarity in response to tyrosine kinase signalling. EMBO J. 2009;28:2360–73.
Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun 2020;11. https://doi.org/10.1038/S41467-020-18794-X.
Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155:1639–51.
Cheung KJ, Padmanaban V, Silvestri V, Schipper K, Cohen JD, Fairchild AN, et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc Natl Acad Sci USA. 2016;113:854.
Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, et al. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51.
Bui NLC, Pandey V, Zhu T, Ma L, Basappa, Lobie PE. Bad phosphorylation as a target of inhibition in oncology. Cancer Lett. 2018;415:177–86.
Kim S, Choi JH, Lim HI, Lee SK, Kim WW, Cho S, et al. EGF-induced MMP-9 expression is mediated by the JAK3/ERK pathway, but not by the JAK3/STAT-3 pathway in a SKBR3 breast cancer cell line. Cell Signal. 2009;21:892–8.
Bodaar K, Yamagata N, Barthe A, Landrigan J, Chonghaile TN, Burns M, et al. JAK3 mutations and mitochondrial apoptosis resistance in T-cell acute lymphoblastic leukemia. Leukemia. 2022;36:1499–507.
Nutter FH, Haylor JL, Khwaja A. Inhibiting ERK activation with CI-1040 leads to compensatory upregulation of alternate MAPKs and Plasminogen activator inhibitor-1 following subtotal nephrectomy with no impact on kidney fibrosis. PLoS One 2015;10. https://doi.org/10.1371/JOURNAL.PONE.0137321.
Allen LF, Sebolt-Leopold J, Meyer MB. CI-1040 (PD184352), a targeted signal transduction inhibitor of MEK (MAPKK). Semin Oncol. 2003;30:105–16.
Li J, Zhao W, Akbani R, Liu W, Ju Z, Ling S, et al. Characterization of human cancer cell lines by reverse-phase protein arrays. Cancer Cell. 2017;31:225–39.
Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503.
Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20.
Wolf DM, Yau C, Wulfkuhle J, Brown-Swigart L, Gallagher IR, Lee PRE, et al. Redefining breast cancer subtypes to guide treatment prioritization and maximize response: Predictive biomarkers across 10 cancer therapies. Cancer Cell. 2022;40:609–23.e6.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Loi S, Dushyanthen S, Beavis PA, Salgado R, Denkert C, Savas P, et al. RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint inhibitors. Clin Cancer Res. 2016;22:1499–509.
Dushyanthen S, Teo ZL, Caramia F, Savas P, Mintoff CP, Virassamy B, et al. Agonist immunotherapy restores T cell function following MEK inhibition improving efficacy in breast cancer. Nat Commun 2017;8. https://doi.org/10.1038/S41467-017-00728-9.
Sastry KSR, Al-Muftah MA, Li P, Al-Kowari MK, Wang E, Ismail Chouchane A, et al. Targeting proapoptotic protein BAD inhibits survival and self-renewal of cancer stem cells. Cell Death Differ. 2014;21:1936–49.
Chu X, Tian W, Ning J, Xiao G, Zhou Y, Wang Z, et al. Cancer stem cells: advances in knowledge and implications for cancer therapy. Signal Transduct Target Ther 2024;9. https://doi.org/10.1038/S41392-024-01851-Y.
Aiello NM, Stanger BZ. Echoes of the embryo: using the developmental biology toolkit to study cancer. Dis Model Mech. 2016;9:105–14.
Cai W, Ye Q, She QB. Loss of 4E-BP1 function induces EMT and promotes cancer cell migration and invasion via cap-dependent translational activation of snail. Oncotarget. 2014;5:6015–27.
Martineau Y, Azar R, Müller D, Lasfargues C, El Khawand S, Anesia R, et al. Pancreatic tumours escape from translational control through 4E-BP1 loss. Oncogene. 2014;33:1367–74.
Wang J, Ye Q, Cao Y, Guo Y, Huang X, Mi W, et al. Snail determines the therapeutic response to mTOR kinase inhibitors by transcriptional repression of 4E-BP1. Nat Commun 2017;8. https://doi.org/10.1038/S41467-017-02243-3.
Taddei I, Deugnier MA, Faraldo MM, Petit V, Bouvard D, Medina D, et al. Beta1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nat Cell Biol. 2008;10:716–22.
Huck L, Pontier SM, Zuo DM, Muller WJ. beta1-integrin is dispensable for the induction of ErbB2 mammary tumors but plays a critical role in the metastatic phase of tumor progression. Proc Natl Acad Sci USA. 2010;107:15559–64.
Johnson MD, Kenney N, Stoica A, Hilakivi-Clarke L, Singh B, Chepko G, et al. Cadmium mimics the in vivo effects of estrogen in the uterus and mammary gland. Nat Med. 2003;9:1081–4.
Parodi DA, Greenfield M, Evans C, Chichura A, Alpaugh A, Williams J, et al. Alteration of mammary gland development and gene expression by in utero exposure to Cadmium. Int J Mol Sci. 2017;18. https://doi.org/10.3390/IJMS18091939.
Wei Z, Shaikh ZA. Cadmium stimulates metastasis-associated phenotype in triple-negative breast cancer cells through integrin and β-catenin signaling. Toxicol Appl Pharm. 2017;328:70–80.
Liang Y, Pi H, Liao L, Tan M, Deng P, Yue Y, et al. Cadmium promotes breast cancer cell proliferation, migration and invasion by inhibiting ACSS2/ATG5-mediated autophagy. Environ Pollut 2021;273. https://doi.org/10.1016/J.ENVPOL.2021.116504.
Coffey DS. Self-organization, complexity and chaos: the new biology for medicine. Nat Med. 1998;4:882–5.
Leth-Larsen R, Lund R, Hansen HV, Laenkholm AV, Tarin D, Jensen ON, et al. Metastasis-related plasma membrane proteins of human breast cancer cells identified by comparative quantitative mass spectrometry. Mol Cell Proteom. 2009;8:1436–49.
Harburg GC, Hinck L. Navigating breast cancer: axon guidance molecules as breast cancer tumor suppressors and oncogenes. J Mammary Gland Biol Neoplasia. 2011;16:257–70.
Brantley-Sieders DM, Jiang A, Sarma K, Badu-Nkansah A, Walter DL, Shyr Y, et al. Eph/ephrin profiling in human breast cancer reveals significant associations between expression level and clinical outcome. PLoS One 2011; 6. https://doi.org/10.1371/JOURNAL.PONE.0024426.
Ogawa K, Pasqualini R, Lindberg RA, Kain R, Freeman AL, Pasquale EB. The ephrin-A1 ligand and its receptor, EphA2, are expressed during tumor neovascularization. Oncogene. 2000;19:6043–52.
Fang WB, Brantley-Sieders DM, Parker MA, Reith AD, Chen J. A kinase-dependent role for EphA2 receptor in promoting tumor growth and metastasis. Oncogene. 2005;24:7859–68.
Brantley-Sieders DM, Zhuang G, Hicks D, Wei BF, Hwang Y, Cates JMM, et al. The receptor tyrosine kinase EphA2 promotes mammary adenocarcinoma tumorigenesis and metastatic progression in mice by amplifying ErbB2 signaling. J Clin Invest. 2008;118:64–78.
Pan M. Overexpression of EphA2 gene in invasive human breast cancer and its association with hormone receptor status. 2005;23:9583.
Mittal V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu Rev Pathol. 2018;13:395–412.
Slaney CY, Möller A, Hertzog PJ, Parker BS. The role of Type I interferons in immunoregulation of breast cancer metastasis to the bone. Oncoimmunology 2013;2. https://doi.org/10.4161/ONCI.22339.
Bidwell BN, Slaney CY, Withana NP, Forster S, Cao Y, Loi S, et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat Med. 2012;18:1224–31.
Siddiqa A, Marciniak R. Targeting the hallmarks of cancer. Cancer Biol Ther. 2008;7:740–1.
VanDyke D, Kyriacopulos P, Yassini B, Wright A, Burkhart E, Jacek S, et al. Nanoparticle Based Combination Treatments for Targeting Multiple Hallmarks of Cancer. Int J Nano Stud Technol 2016;1–18.
Luo Y, Wu Y, Huang H, Yi N, Chen Y. Emerging role of BAD and DAD1 as potential targets and biomarkers in cancer. Oncol Lett 2021;22. https://doi.org/10.3892/OL.2021.13072.
Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–39.
Chattopadhyay A, Chiang CW, Yang E. BAD/BCL-[X(L)] heterodimerization leads to bypass of G0/G1 arrest. Oncogene. 2001;20:4507–18.
Danial NN, Gramm CF, Scorrano L, Zhang C-Y, Krauss S, Ranger AM, et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–6.
Giménez-Cassina A, Garcia-Haro L, Choi CS, Osundiji MA, Lane EA, Huang H, et al. Regulation of hepatic energy metabolism and gluconeogenesis by BAD. Cell Metab. 2014;19:272–84.
Danial NN, Walensky LD, Zhang CY, Choi CS, Fisher JK, Molina AJ, et al. Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat Med. 2008;14:144–53.
Githaka JM, Gutiérrez T, Han ZZ, Primeau JO, Muranyi H, Young HS, et al. The non-cytotoxic small molecule NPB does not inhibit BAD phosphorylation and forms colloidal aggregates. Commun Med. (In Press) 2025. https://doi.org/10.1038/s43856-025-00880-0.
Gilmore AP, Valentijn AJ, Wang P, Ranger AM, Bundred N, O’Hare MJ, et al. Activation of BAD by therapeutic inhibition of epidermal growth factor receptor and transactivation by insulin-like growth factor receptor. J Biol Chem. 2002;277:27643–50.
Scheid MP, Schubert KM, Duronio V. Regulation of bad phosphorylation and association with Bcl-x(L) by the MAPK/Erk kinase. J Biol Chem. 1999;274:31108–13.
Guo Y, Pan W, Liu S, Shen Z, Xu Y, Hu L. ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med 2020;19. https://doi.org/10.3892/ETM.2020.8454.
Acosta-Casique A, Montes-Alvarado JB, Barragán M, Larrauri-Rodríguez KA, Perez-Gonzalez A, Delgado-Magallón A, et al. ERK activation modulates invasiveness and Reactive Oxygen Species (ROS) production in triple negative breast cancer cell lines. Cell Signal 2023;101. https://doi.org/10.1016/J.CELLSIG.2022.110487.
Bartholomeusz C, Gonzalez-Angulo AM, Liu P, Hayashi N, Lluch A, Ferrer-Lozano J, et al. High ERK protein expression levels correlate with shorter survival in triple-negative breast cancer patients. Oncologist. 2012;17:766–74.
Zhang Y, Lu Q, Li N, Xu M, Miyamoto T, Liu J. Sulforaphane suppresses metastasis of triple-negative breast cancer cells by targeting the RAF/MEK/ERK pathway. NPJ Breast Cancer 2022;8. https://doi.org/10.1038/S41523-022-00402-4.
Gagliardi M, Pitner MK, Park J, Xie X, Saso H, Larson RA, et al. Differential functions of ERK1 and ERK2 in lung metastasis processes in triple-negative breast cancer. Sci Rep 2020;10. https://doi.org/10.1038/S41598-020-65250-3.
Rinehart J, Adjei AA, LoRusso PM, Waterhouse D, Hecht JR, Natale RB, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol. 2004;22:4456–62.
Boasberg PD, Redfern CH, Daniels GA, Bodkin D, Garrett CR, Ricart AD. Pilot study of PD-0325901 in previously treated patients with advanced melanoma, breast cancer, and colon cancer. Cancer Chemother Pharm. 2011;68:547–52.
LoRusso PM, Krishnamurthi SS, Rinehart JJ, Nabell LM, Malburg L, Chapman PB, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral MAPK/ERK kinase inhibitor PD-0325901 in patients with advanced cancers. Clin Cancer Res. 2010;16:1924–37.
Cheng Y, Tian H. Current Development Status of MEK Inhibitors. Molecules 2017; 22. https://doi.org/10.3390/MOLECULES22101551.
Acknowledgements
We are grateful to all Goping lab members for their valuable discussions and contributions. We thank Dr. Kristi Baker for the generous gift of anti-CD8 antibody. Cell Imaging Core Experiments were performed at the University of Alberta Faculty of Medicine & Dentistry Cell Imaging Core, RRID:SCR_019200, which receives financial support from the Faculty of Medicine & Dentistry, the University Hospital Foundation, Striving for Pandemic Preparedness – The Alberta Research Consortium, and Canada Foundation for Innovation (CFI) awards to contributing investigators. The authors acknowledge the dedicated support and expertise provided by the North Campus Animal Services (NCAS) staff and veterinarians. This work was supported by operating grants from the Alberta Cancer Foundation and Canadian Institutes of Health Research to ISG. ISG gratefully acknowledges support as the Lilian McCullough Chair in Breast Cancer Research.
Author information
Authors and Affiliations
Contributions
I.S.G. conceived the study. J.M.G., R.K., N.P., and I.S.G. designed the research. J.M.G., R.K., N.T., J.W., H.M., N.T., L.P., E.P.L., T.P., and R.M. performed the experiments. N.D. provided mouse strains. J.M.G., N.P., P.N.N., and J.W. analyzed the data. I.S.G. directed research. J.M.G. and I.S.G. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All mouse experiments and procedures were performed in accordance with the guidelines and regulations set forth by the Canadian Council on Animal Care and approved by the University of Alberta Health Sciences Animal Care and Use Committee (Protocol# AUP00000386).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Githaka, J.M., Kirschenman, R., Patel, N. et al. Multiple anti-tumor programs are activated by blocking BAD phosphorylation. Oncogene 44, 2530–2546 (2025). https://doi.org/10.1038/s41388-025-03420-1
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03420-1