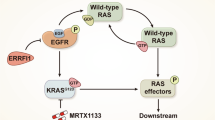
Abstract
The ERK1/2 signaling pathway, one of the most frequently dysregulated oncogenic pathways, can be initiated by diverse mutations, including those in RAS, BRAF, and amplifications of ERBB2 (HER2). Co-occurrence of ERK1/2 hyperactivation and TP53 mutations is common in multiple cancer types and correlates with significantly poorer clinical outcomes. However, the direct mechanisms underlying the cooperation between ERK1/2 signaling and mutant p53 remain largely unexplored. Our study demonstrates that oncogenic KRAS activates c-JUN, which facilitates physical interactions with mutant p53, leading to hyperactivation of several pro-metastatic transcriptional networks. Notably, mutant p53 and c-JUN collaboratively upregulate EGR1, a key driver of tumor invasion and metastasis. The combined effects of elevated EGR1 expression, along with signaling pathways activated by KRAS and mutant p53, significantly enhance pro-metastatic traits in cancer cells. These findings provide crucial insights into the co-enrichment of KRAS and p53 mutations and pave the way for novel therapeutic strategies targeting this interaction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The RNA sequencing data generated from mutant p53 knockdown experiments in this study are available in the GEO Repository under accession numbers GSE65458, GSE68248, and GSE291033. For ChIP-seq data: MDA-MB-231 cell line: p53 and c-JUN data are accessible through GEO (GSE66543, GSE66081). A549 cell line: p53 (ENCFF877LNP) and c-JUN (ENCFF957TUK) data are available via ENCODE. HepG2 cell line: p53 and c-JUN data can be found in ENCODE under accession number ENCFF784KMC (p53) and ENCFF732BEG(c-JUN).
Code availability
The custom code and scripts used for data analysis in this study are available from the corresponding author upon reasonable request. All software packages and publicly available tools utilized in this work are cited in the Methods section, along with their respective version numbers and accessibility information.
References
Maik-Rachline G, Hacohen-Lev-Ran A, Seger R. Nuclear ERK: mechanism of translocation, substrates, and role in cancer. Int J Mol Sci. 2019;20:1194.
Lavoie H, Gagnon J, Therrien M. ERK signalling: a master regulator of cell behaviour, life and fate. Nat Rev Mol Cell Biol. 2020;21:607–32.
Martinez-Jimenez F, Muinos F, Sentis I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20:555–72.
Prior IA, Hood FE, Hartley JL. The frequency of Ras mutations in cancer. Cancer Res. 2020;80:2969–74.
Muzumdar MD, Dorans KJ, Chung KM, Robbins R, Tammela T, Gocheva V, et al. Clonal dynamics following p53 loss of heterozygosity in Kras-driven cancers. Nat Commun. 2016;7:12685.
Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129:1287–92.
Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–67.
Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33.
Leroy B, Anderson M, Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat. 2014;35:672–88.
Chun YS, Passot G, Yamashita S, Nusrat M, Katsonis P, Loree JM, et al. Deleterious effect of RAS and evolutionary high-risk TP53 double mutation in colorectal liver metastases. Ann Surg. 2019;269:917–23.
Du L, Kim JJ, Shen J, Chen B, Dai N. KRAS and TP53 mutations in inflammatory bowel disease-associated colorectal cancer: a meta-analysis. Oncotarget. 2017;8:22175–86.
Zhao J, Han Y, Li J, Chai R, Bai C. Prognostic value of KRAS/TP53/PIK3CA in non-small cell lung cancer. Oncol Lett. 2019;17:3233–40.
Escobar-Hoyos LF, Penson A, Kannan R, Cho H, Pan CH, Singh RK, et al. Altered RNA splicing by mutant p53 activates oncogenic RAS signaling in pancreatic cancer. Cancer Cell. 2020;38:198–211 e8.
Kim MP, Li X, Deng J, Zhang Y, Dai B, Allton KL, et al. Oncogenic KRAS recruits an expansive transcriptional network through mutant p53 to drive pancreatic cancer metastasis. Cancer Discov. 2021;11:2094–111.
Silwal-Pandit L, Langerod A, Borresen-Dale AL. TP53 mutations in breast and ovarian cancer. Cold Spring Harb Perspect Med. 2017;7:a026252.
Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35:600–4.
DiNatale A, Castelli MS, Nash B, Meucci O, Fatatis A. Regulation of tumor and metastasis initiation by chemokine receptors. J Cancer. 2022;13:3160–76.
Tanimura S, Takeda K. ERK signalling as a regulator of cell motility. J Biochem. 2017;162:145–54.
Samson SC, Khan AM, Mendoza MC. ERK signaling for cell migration and invasion. Front Mol Biosci. 2022;9:998475.
Yue X, Zhao Y, Xu Y, Zheng M, Feng Z, Hu W. Mutant p53 in cancer: accumulation, gain-of-function, and therapy. J Mol Biol. 2017;429:1595–606.
Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–18.
Yang L, Zheng L, Chng WJ, Ding JL. Comprehensive analysis of ERK1/2 substrates for potential combination immunotherapies. Trends Pharm Sci. 2019;40:897–910.
Li TT, Liu MR, Pei DS. Friend or foe, the role of EGR-1 in cancer. Med Oncol. 2019;37:7.
Sun T, Tian H, Feng YG, Zhu YQ, Zhang WQ. Egr-1 promotes cell proliferation and invasion by increasing beta-catenin expression in gastric cancer. Dig Dis Sci. 2013;58:423–30.
Hoffmann E, Ashouri J, Wolter S, Doerrie A, Dittrich-Breiholz O, Schneider H, et al. Transcriptional regulation of EGR-1 by the interleukin-1-JNK-MKK7-c-Jun pathway. J Biol Chem. 2008;283:12120–8.
Li Y, Samuvel DJ, Sundararaj KP, Lopes-Virella MF, Huang Y. IL-6 and high glucose synergistically upregulate MMP-1 expression by U937 mononuclear phagocytes via ERK1/2 and JNK pathways and c-Jun. J Cell Biochem. 2010;110:248–59.
Chakraborty A, Diefenbacher ME, Mylona A, Kassel O, Behrens A. The E3 ubiquitin ligase Trim7 mediates c-Jun/AP-1 activation by Ras signalling. Nat Commun. 2015;6:6782.
Hu S, Wang M, Ji A, Yang J, Gao R, Li X, et al. Mutant p53 and ELK1 co-drive FRA-1 expression to induce metastasis in breast cancer. FEBS Lett. 2023;597:3087–101.
Adiseshaiah P, Li J, Vaz M, Kalvakolanu DV, Reddy SP. ERK signaling regulates tumor promoter induced c-Jun recruitment at the Fra-1 promoter. Biochem Biophys Res Commun. 2008;371:304–8.
Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–86.
Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8.
Zhang C, Liu J, Xu D, Zhang T, Hu W, Feng Z. Gain-of-function mutant p53 in cancer progression and therapy. J Mol Cell Biol. 2020;12:674–87.
Pfister NT, Prives C. Transcriptional regulation by wild-type and cancer-related mutant forms of p53. Cold Spring Harb Perspect Med. 2017;7:a026054.
Wang B, Guo H, Yu H, Chen Y, Xu H, Zhao G. The role of the transcription factor EGR1 in cancer. Front Oncol. 2021;11:642547.
Acknowledgements
This work was supported Science and Technology Fund of Tianjin Municipal Health Commission (NO. TJWJ2022QN098).
Author information
Authors and Affiliations
Contributions
MXW and ALJ performed the experiments and data analysis. RFG performed data analysis. SKH designed the study and wrote the manuscript. MXW acquired the funding. All authors edited and revised the manuscript and were involved in the final approval of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study did not involve human participants, human tissue, or live vertebrate animals. All analyses were conducted using publicly available datasets and computational modeling techniques. Therefore, ethical approval from our institutional ethics committee and informed consent were not required for this research.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, M., Ji, A., Gao, R. et al. Coexistence of P53 and KRAS mutations enhances ERK1/2 signaling by inducing EGR1 expression through mutp53 and c-JUN interaction. Oncogene 44, 3787–3798 (2025). https://doi.org/10.1038/s41388-025-03536-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03536-4