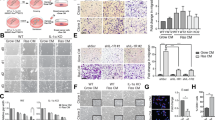
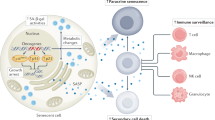
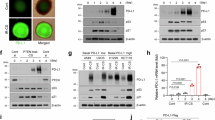
Abstract
Triple-negative breast cancer (TNBC) is the most aggressive form of breast cancer that is associated with poor prognosis and a high risk of relapse, with limited treatment options. While the induction of senescence, a state of arrested cell growth, is generally achieved by available anticancer treatments, senescence can adversely promote tumorigenesis through an upheld augmented inflammatory state called senescence-associated secretory phenotype (SASP). Thus, the precise delineation of underlying regulatory mechanisms governing senescence is urgently needed. Herein, we investigated the beneficial anticancer senescence response elicited by silencing the expression of the promyelocytic leukemia protein (PML) in TNBC, where it exerts an oncogenic role. Functional genomics studies implicated the downregulation of a specific set of ribosomal protein (RP) genes tied to poor clinical outcome. Re-introduction of RPL38 or RPL39L alone, but not RPS14, a favorable outcome-associated RP, was sufficient to block the senescence phenotype induced by PML knockdown. RP gene regulation by PML was found to involve the assembly of a previously unrecognized PML-mTOR-RONIN transcriptional complex at their promoters. Furthermore, we show that RONIN levels are elevated in TNBC and that RONIN silencing can recapitulate the senescent phenotype of PML-deficient cells. This work offers new therapeutic insights for TNBC that involve senescence-inducing therapies or senolytics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
PML and mTOR ChIP-seq data performed in MDA-MB-231 cells, as well as RNA-seq data of shRNA-mediated knockdown of PML in MDA-MB-231 cells, have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO SuperSeries accession number GSE283109 encompassing SubSeries GSE283106 (shPML RNA-seq), GSE283107 (PML ChIP-seq), and GSE283108 (mTOR ChIP-seq). Gene expression data from the breast cancer METABRIC [63, 64] and TCGA PanCancer atlas cohorts were downloaded from cbioportal (https://www.cbioportal.org) [56, 57] or analyzed with bc-GenExMiner v5.1 (https://bcgenex.ico.unicancer.fr/BC-GEM/). Proteomics data from a breast cancer patient cohort were obtained from CPTAC [59]. Proteomics data of breast cancer cell lines were obtained from (1) CCLE [60] via the Dependency Map (DepMap) portal (https://depmap.org/portal/); (2) Lapek et al. [61]; or (3) Sun et al. [62]. This paper does not report original code. Source data underlying the graphs are presented in Supplementary Table 8. Uncropped immunoblots are shown in Supplementary Fig. S6. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
Al-Thoubaity FK. Molecular classification of breast cancer: a retrospective cohort study. Ann Med Surg. 2020;49:44–8.
de Paula B, Kieran R, Koh SSY, Crocamo S, Abdelhay E, Munoz-Espin D. Targeting senescence as a therapeutic opportunity for triple-negative breast cancer. Mol Cancer Ther. 2023;22:583–98.
Schmitt CA, Wang B, Demaria M. Senescence and cancer—role and therapeutic opportunities. Nat Rev Clin Oncol. 2022;19:619–36.
Bousset L, Gil J. Targeting senescence as an anticancer therapy. Mol Oncol. 2022;16:3855–80.
Wang L, Lankhorst L, Bernards R. Exploiting senescence for the treatment of cancer. Nat Rev Cancer. 2022;22:340–55.
Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of cellular senescence. Trends Cell Biol. 2018;28:436–53.
Wang B, Han J, Elisseeff JH, Demaria M. The senescence-associated secretory phenotype and its physiological and pathological implications. Nat Rev Mol Cell Biol. 2024;25:958–78.
Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–7.
Demaria M, O’Leary MN, Chang J, Shao L, Liu S, Alimirah F, et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 2017;7:165–76.
Xiao S, Qin D, Hou X, Tian L, Yu Y, Zhang R, et al. Cellular senescence: a double-edged sword in cancer therapy. Front Oncol. 2023;13:1189015.
Corpet A, Kleijwegt C, Roubille S, Juillard F, Jacquet K, Texier P, et al. PML nuclear bodies and chromatin dynamics: catch me if you can!. Nucleic Acids Res. 2020;48:11890–912.
Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW. PML is induced by oncogenic Ras and promotes premature senescence. Genes Dev. 2000;14:2015–27.
Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18:1350–8.
Tessier S, Martin-Martin N, de The H, Carracedo A, Lallemand-Breitenbach V. Promyelocytic leukemia protein, a protein at the crossroad of oxidative stress and metabolism. Antioxid Redox Signal. 2017;26:432–44.
Wang ZG, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, et al. PML is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–72.
Vernier M, Bourdeau V, Gaumont-Leclerc MF, Moiseeva O, Begin V, Saad F, et al. Regulation of E2Fs and senescence by PML nuclear bodies. Genes Dev. 2011;25:41–50.
Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–16.
Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, et al. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst. 2004;96:269–79.
Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cardon-Cardo- C, et al. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–51.
Mazza M, Pelicci PG. Is PML a tumor suppressor?. Front Oncol. 2013;3:174.
Carracedo A, Weiss D, Leliaert AK, Bhasin M, de Boer VC, Laurent G, et al. A metabolic prosurvival role for PML in breast cancer. J Clin Investig. 2012;122:3088–100.
Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, et al. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453:1072–8.
Liu SB, Shen ZF, Guo YJ, Cao LX, Xu Y. PML silencing inhibits cell proliferation and induces DNA damage in cultured ovarian cancer cells. Biomed Rep. 2017;7:29–35.
Zhou W, Cheng L, Shi Y, Ke SQ, Huang Z, Fang X, et al. Arsenic trioxide disrupts glioma stem cells via promoting PML degradation to inhibit tumor growth. Oncotarget. 2015;6:37300–15.
Martin-Martin N, Piva M, Urosevic J, Aldaz P, Sutherland JD, Fernandez-Ruiz S, et al. Stratification and therapeutic potential of PML in metastatic breast cancer. Nat Commun. 2016;7:12595.
Ponente M, Campanini L, Cuttano R, Piunti A, Delledonne GA, Coltella N, et al. PML promotes metastasis of triple-negative breast cancer through transcriptional regulation of HIF1A target genes. JCI Insight. 2017;2:e87380.
Arreal L, Piva M, Fernandez S, Revandkar A, Schaub-Clerigue A, Villanueva J, et al. Targeting PML in triple negative breast cancer elicits growth suppression and senescence. Cell Death Differ. 2020;27:1186–99.
Bringold F, Serrano M. Tumor suppressors and oncogenes in cellular senescence. Exp Gerontol. 2000;35:317–29.
Peng Y, Wang Y, Zhou C, Mei W, Zeng C. PI3K/Akt/mTOR pathway and its role in cancer therapeutics: Are we making headway?. Front Oncol. 2022;12:819128.
Tian T, Li X, Zhang J. mTOR signaling in cancer and mTOR inhibitors in solid tumor targeting therapy. Int J Mol Sci. 2019;20:755.
Glaviano A, Foo ASC, Lam HY, Yap KCH, Jacot W, Jones RH, et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer. 2023;22:138.
Miricescu D, Totan A, Stanescu S II, Badoiu SC, Stefani C, Greabu M. PI3K/AKT/mTOR signaling pathway in breast cancer: from molecular landscape to clinical aspects. Int J Mol Sci. 2020;22:173.
Popova NV, Jucker M. The role of mTOR signaling as a therapeutic target in cancer. Int J Mol Sci. 2021;22:1743.
Giguère V. Canonical signaling and nuclear activity of mTOR—a teamwork effort to regulate metabolism and cell growth. Febs J. 2018;285:1572–88.
Laribee RN, Weisman R. Nuclear functions of TOR: impact on transcription and the epigenome. Genes. 2020;11:641.
Panwar V, Singh A, Bhatt M, Tonk RK, Azizov S, Raza AS, et al. Multifaceted role of mTOR (mammalian target of rapamycin) signaling pathway in human health and disease. Signal Transduct Target Ther. 2023;8:375.
Simcox J, Lamming DW. The central moTOR of metabolism. Dev Cell. 2022;57:691–706.
Demidenko ZN, Zubova SG, Bukreeva EI, Pospelov VA, Pospelova TV, Blagosklonny MV. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8:1888–95.
Kolesnichenko M, Hong L, Liao R, Vogt PK, Sun P. Attenuation of TORC1 signaling delays replicative and oncogenic RAS-induced senescence. Cell Cycle. 2012;11:2391–401.
Leontieva OV, Blagosklonny MV. DNA damaging agents and p53 do not cause senescence in quiescent cells, while consecutive re-activation of mTOR is associated with conversion to senescence. Aging. 2010;2:924–35.
Leontieva OV, Blagosklonny MV. Gerosuppression by pan-mTOR inhibitors. Aging. 2016;8:3535–51.
Pospelova TV, Leontieva OV, Bykova TV, Zubova SG, Pospelov VA, Blagosklonny MV. Suppression of replicative senescence by rapamycin in rodent embryonic cells. Cell Cycle. 2012;11:2402–7.
Xu S, Cai Y, Wei Y. mTOR signaling from cellular senescence to organismal aging. Aging Dis. 2014;5:263–73.
Cayo A, Segovia R, Venturini W, Moore-Carrasco R, Valenzuela C, Brown N. mTOR activity and autophagy in senescent cells, a complex partnership. Int J Mol Sci. 2021;22:8149.
Alimonti A, Nardella C, Chen Z, Clohessy JG, Carracedo A, Trotman LC, et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Investig. 2010;120:681–93.
Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332:966–70.
Laberge RM, Sun Y, Orjalo AV, Patil CK, Freund A, Zhou L, et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol. 2015;17:1049–61.
Herranz N, Gallage S, Mellone M, Wuestefeld T, Klotz S, Hanley CJ, et al. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat Cell Biol. 2015;17:1205–17.
Liu Y, Azizian NG, Sullivan DK, Li Y. mTOR inhibition attenuates chemosensitivity through the induction of chemotherapy resistant persisters. Nat Commun. 2022;13:7047.
Vilotti S, Biagioli M, Foti R, Dal Ferro M, Lavina ZS, Collavin L, et al. The PML nuclear bodies-associated protein TTRAP regulates ribosome biogenesis in nucleolar cavities upon proteasome inhibition. Cell Death Differ. 2012;19:488–500.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89.
Khan A, Mathelier A. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinform. 2017;18:287.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–45.
Krug K, Jaehnig EJ, Satpathy S, Blumenberg L, Karpova A, Anurag M, et al. Proteogenomic landscape of breast cancer tumorigenesis and targeted therapy. Cell. 2020;183:1436–56.e31.
Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER 3rd, et al. Quantitative proteomics of the cancer cell line encyclopedia. Cell. 2020;180:387–402.e16.
Lapek JD Jr., Greninger P, Morris R, Amzallag A, Pruteanu-Malinici I, Benes CH, et al. Detection of dysregulated protein-association networks by high-throughput proteomics predicts cancer vulnerabilities. Nat Biotechnol. 2017;35:983–9.
Sun R, Ge W, Zhu Y, Sayad A, Luna A, Lyu M, et al. Proteomic dynamics of breast cancer cell lines identifies potential therapeutic protein targets. Mol Cell Proteom. 2023;22:100602.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52.
Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479.
Jochems F, Thijssen B, De Conti G, Jansen R, Pogacar Z, Groot K, et al. The Cancer SENESCopedia: a delineation of cancer cell senescence. Cell Rep. 2021;36:109441.
Liu Y, Deisenroth C, Zhang Y. RP-MDM2-p53 pathway: linking ribosomal biogenesis and tumor surveillance. Trends Cancer. 2016;2:191–204.
Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–17.
Bernardi R, Guernah I, Jin D, Grisendi S, Alimonti A, Teruya-Feldstein J, et al. PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR. Nature. 2006;442:779–85.
Audet-Walsh E, Dufour CR, Yee T, Zouanat FZ, Yan M, Kalloghlian G, et al. Nuclear mTOR acts as a transcriptional integrator of the androgen signaling pathway in prostate cancer. Genes Dev. 2017;31:1228–42.
Chaveroux C, Eichner LJ, Dufour CR, Shatnawi A, Khoutorsky A, Bourque G, et al. Molecular and genetic crosstalks between mTOR and ERRalpha are key determinants of rapamycin-induced nonalcoholic fatty liver. Cell Metab. 2013;17:586–98.
Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–40.
Dufour CR, Scholtes C, Yan M, Chen Y, Han L, Li T, et al. The mTOR chromatin-bound interactome in prostate cancer. Cell Rep. 2022;38:110534.
Sabogal A, Lyubimov AY, Corn JE, Berger JM, Rio DC. THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves. Nature Struct Mol Biol. 2010;17:117–23.
Sanghavi HM, Mallajosyula SS, Majumdar S. Classification of the human THAP protein family identifies an evolutionarily conserved coiled coil region. BMC Struct Biol. 2019;19:4.
Fujita J, Freire P, Coarfa C, Benham AL, Gunaratne P, Schneider MD, et al. Ronin governs early heart development by controlling core gene expression programs. Cell Rep. 2017;21:1562–73.
Seifert BA, Dejosez M, Zwaka TP. Ronin influences the DNA damage response in pluripotent stem cells. Stem Cell Res. 2017;23:98–104.
Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, et al. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–74.
Lian WX, Yin RH, Kong XZ, Zhang T, Huang XH, Zheng WW, et al. THAP11, a novel binding protein of PCBP1, negatively regulates CD44 alternative splicing and cell invasion in a human hepatoma cell line. FEBS Lett. 2012;586:1431–8.
Nakamura S, Yokota D, Tan L, Nagata Y, Takemura T, Hirano I, et al. Down-regulation of Thanatos-associated protein 11 by BCR-ABL promotes CML cell proliferation through c-Myc expression. Int J Cancer. 2012;130:1046–59.
Parker JB, Palchaudhuri S, Yin H, Wei J, Chakravarti D. A transcriptional regulatory role of the THAP11-HCF-1 complex in colon cancer cell function. Mol Cell Biol. 2012;32:1654–70.
Zhang J, Zhang H, Shi H, Wang F, Du J, Wang Y, et al. THAP11 Functions as a tumor suppressor in gastric cancer through regulating c-Myc signaling pathways. Biomed Res Int. 2020;2020:7838924.
Lessard F, Igelmann S, Trahan C, Huot G, Saint-Germain E, Mignacca L, et al. Senescence-associated ribosome biogenesis defects contributes to cell cycle arrest through the Rb pathway. Nat Cell Biol. 2018;20:789–99.
Li HY, Wang M, Jiang X, Jing Y, Wu Z, He Y, et al. CRISPR screening uncovers nucleolar RPL22 as a heterochromatin destabilizer and senescence driver. Nucleic Acids Res. 2024;52:11481–99.
Artero-Castro A, Kondoh H, Fernandez-Marcos PJ, Serrano M, Ramon y Cajal S, Lleonart ME. Rplp1 bypasses replicative senescence and contributes to transformation. Exp Cell Res. 2009;315:1372–83.
Pecoraro A, Pagano M, Russo G, Russo A. Ribosome biogenesis and cancer: overview on ribosomal proteins. Int J Mol Sci. 2021;22:5496.
Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21:164–5.
Jiao L, Liu Y, Yu XY, Pan X, Zhang Y, Tu J, et al. Ribosome biogenesis in disease: new players and therapeutic targets. Signal Transduct Target Ther. 2023;8:15.
Chern T, Achilleos A, Tong X, Hill MC, Saltzman AB, Reineke LC, et al. Mutations in Hcfc1 and Ronin result in an inborn error of cobalamin metabolism and ribosomopathy. Nat Commun. 2022;13:134.
Hsu KS, Kao HY. PML: Regulation and multifaceted function beyond tumor suppression. Cell Biosci. 2018;8:5.
Fracassi C, Ugge M, Abdelhalim M, Zapparoli E, Simoni M, Magliulo D, et al. PML modulates epigenetic composition of chromatin to regulate expression of pro-metastatic genes in triple-negative breast cancer. Nucleic Acids Res. 2023;51:11024–39.
Mijit M, Caracciolo V, Melillo A, Amicarelli F, Giordano A. Role of p53 in the regulation of cellular senescence. Biomolecules. 2020;10:420.
Simoni M, Menegazzi C, Fracassi C, Biffi CC, Genova F, Tenace NP, et al. PML restrains p53 activity and cellular senescence in clear cell renal cell carcinoma. EMBO Mol Med. 2024;16:1324–51.
Chen X, Zhang T, Su W, Dou Z, Zhao D, Jin X, et al. Mutant p53 in cancer: from molecular mechanism to therapeutic modulation. Cell Death Dis. 2022;13:974.
Haupt S, di Agostino S, Mizrahi I, Alsheich-Bartok O, Voorhoeve M, Damalas A, et al. Promyelocytic leukemia protein is required for gain of function by mutant p53. Cancer Res. 2009;69:4818–26.
Acknowledgements
The authors thank members of the Giguère laboratory for assistance and critical discussions, the McGill Platform for Cellular Perturbation (MPCP), and Mr. Alain Pacis from the Canadian Centre for Computational Genomics (C3G) for help with ChIP-seq analysis. Results from examination of breast cancer mRNA-sequencing data from the TCGA-pan cancer dataset are based upon data generated by the TCGA Research Network (https://www.cancer.gov/tcga). This work was supported by a Foundation grant from the Canadian Institutes of Health Research (CIHR) to VG (FDT-156254), a Terry Fox Research Institute Team Grant (PPG-1091), and an operating grant from the Cancer Research Society (CRS-1052927). YM is supported by a Charlotte & Leo Karassik Fellowship and a FRQS Postdoctoral Fellowship. LH is supported by Canderel and Fond de recherches du Québec – Santé (FRQS) studentships. AA is supported by a CIHR studentship. PH is supported by a J.P. Collip fellowship in medical research from McGill’s Faculty of Medicine and Health Sciences (FMHS). MV was a recipient of a post-doctoral fellowship from CIHR. AR was supported by a Canderel post-doctoral fellowship.
Author information
Authors and Affiliations
Contributions
Conceptualization: M Vernier, V Giguère, and CR Dufour; methodology: M Vernier, Y Medkour; investigation: Y Medkour, CR Dufour, L Han, P Hutton, M Farhat, A Alfonso, A Rambur, M Vernier, and V Giguère; writing—original draft: M Vernier, CR Dufour; Y Medkour; writing—review and editing: V Giguère; study supervision: V Giguère and M Vernier; funding acquisition: V Giguère.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Medkour, Y., Dufour, C.R., Han, L. et al. Regulation of ribosomal gene expression and senescence by a PML-mTOR-RONIN nuclear complex in triple-negative breast cancer. Oncogene 44, 4712–4726 (2025). https://doi.org/10.1038/s41388-025-03623-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03623-6