Abstract
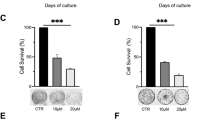
The Polycomb group (PcG) protein chromobox 8 (CBX8) is the subunit of Polycomb repressive complex 1 (PRC1) and recognizes the trimethylation of histone H3 on Lysine 27 (H3K27me3), and coordinates with PRC2 complex to function as an epigenetic gene silencer. CBX8 plays a key role in cell proliferation, stem cell biology, cell senescence, and cancer development. However, the post-translational modifications of CBX8 remain poorly understood. Here, we report that protein kinase D1 (PKD1) interacts and phosphorylates CBX8 at Thr234 and Ser256 /311 residues. PKD1-mediated CBX8 phosphorylation at Thr234 reduced its expression level by promoting its ubiquitination-mediated degradation, whereas Ser256/311 phosphorylation decreased CBX8 binding to other PRC1 components BMI1 and RING1A. Overall, CBX8 phosphorylation by PKD1 impaired PRC1 complex integrity and activity, mitigated H2AK119ub1 level, caused the upregulation of multiple target genes repressed by CBX8, and decreased CBX8, H2AK119ub1, and H3K27me3 enrichment at INK4A/ARF locus, thereby derepressing p16INK4A and facilitating cellular senescence. Collectively, these results suggest that PKD1-mediated CBX8 phosphorylation at T234 and S256/311 is a key mechanism governing CBX8 function, including cell senescence.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD045225. The website is: http://www.ebi.ac.uk/pride.
References
Salama R, Sadaie M, Hoare M, Narita M. Cellular senescence and its effector programs. Genes Dev. 2014;28:99–114. https://doi.org/10.1101/gad.235184.113.
He S, Sharpless NE. Senescence in health and disease. Cell. 2017;169:1000–11. https://doi.org/10.1016/j.cell.2017.05.015.
Muñoz-Espín D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15:482–96. https://doi.org/10.1038/nrm3823.
Gil J, Peters G. Regulation of the INK4b–ARF–INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–77. https://doi.org/10.1038/nrm1987.
Blackledge NP, Klose RJ. The molecular principles of gene regulation by Polycomb repressive complexes. Nat Rev Mol Cell Biol. 2021;22:815–33. https://doi.org/10.1038/s41580-021-00401-1.
Guo Y, Zhao S, Wang GG. Polycomb gene silencing mechanisms: PRC2 chromatin targeting, H3K27me3 ‘readout’, and phase separation-based compaction. Trends Genet. 2021;37:547–65. https://doi.org/10.1016/j.tig.2021.03.005.
Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. https://doi.org/10.1126/science.1076997.
Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8. https://doi.org/10.1038/nature02985.
Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–54. https://doi.org/10.1016/j.molcel.2005.12.002.
Barbour H, Daou S, Hendzel M, Affar el B. Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes. Nat Commun. 2020;11:5947. https://doi.org/10.1038/s41467-020-19782-w.
Kim JJ, Kingston RE. Context-specific Polycomb mechanisms in development. Nat Rev Genet. 2022;23:680–95. https://doi.org/10.1038/s41576-022-00415-1.
Chan HL, Morey L. Emerging roles for Polycomb-group proteins in stem cells and cancer. Trends Biochem Sci. 2019;44:688–700. https://doi.org/10.1016/j.tibs.2019.04.005.
Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–8. https://doi.org/10.1038/16476.
Gil J, Bernard D, Martínez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. https://doi.org/10.1038/ncb1075.
Dietrich N, Bracken AP, Trinh E, Schjerling CK, Koseki H, Rappsilber J, et al. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J. 2007;26:1637–48. https://doi.org/10.1038/sj.emboj.7601618.
Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–30. https://doi.org/10.1101/gad.415507.
Maertens GN, El Messaoudi-Aubert S, Racek T, Stock JK, Nicholls J, Rodriguez-Niedenführ M, et al. Several distinct polycomb complexes regulate and co-localize on the INK4a tumor suppressor locus. PLoS ONE. 2009;4:e6380. https://doi.org/10.1371/journal.pone.0006380.
Voncken JW, Niessen H, Neufeld B, Rennefahrt U, Dahlmans V, Kubben N, et al. MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the polycomb group protein Bmi1. J Biol Chem. 2005;280:5178–87. https://doi.org/10.1074/jbc.M410819200.
Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol. 2010;12:1108–14. https://doi.org/10.1038/ncb2116.
Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–20. https://doi.org/10.1101/gad.1983810.
Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol. 2011;13:87–94. https://doi.org/10.1038/ncb2139.
Wu SC, Zhang Y. Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. J Biol Chem. 2011;286:28511–9. https://doi.org/10.1074/jbc.M111.248377.
Wan L, Xu K, Wei Y, Zhang J, Han T, Fry C, et al. Phosphorylation of EZH2 by AMPK suppresses PRC2 methyltransferase activity and oncogenic function. Mol Cell. 2018;69:279–91.e5. https://doi.org/10.1016/j.molcel.2017.12.020.
Liu Y, Liu F, Yu H, Zhao X, Sashida G, Deblasio A, et al. Akt phosphorylates the transcriptional repressor bmi1 to block its effects on the tumor-suppressing ink4a-arf locus. Sci Signal. 2012;5:ra77. https://doi.org/10.1126/scisignal.2002961.
Wu HA, Balsbaugh JL, Chandler H, Georgilis A, Zullow H, Shabanowitz J, et al. Mitogen-activated protein kinase signaling mediates phosphorylation of polycomb ortholog Cbx7. J Biol Chem. 2013;288:36398–408. https://doi.org/10.1074/jbc.M113.501720.
Kawaguchi T, Machida S, Kurumizaka H, Tagami H, Nakayama JI. Phosphorylation of CBX2 controls its nucleosome-binding specificity. J Biochem. 2017;162:343–55. https://doi.org/10.1093/jb/mvx045.
Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ, et al. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res. 2013;12:260–71. https://doi.org/10.1021/pr300630k.
Zhan X, Yang J, Mao Z, Yu W. PIM1-catalyzed CBX8 phosphorylation promotes the oncogene-induced senescence of human diploid fibroblast. Biochem Biophys Res Commun. 2018;501:779–85. https://doi.org/10.1016/j.bbrc.2018.05.067.
Kim HJ, Park JW, Kang JY, Seo SB. Negative Regulation of Erythroid Differentiation via the CBX8-TRIM28 Axis. Mol Cells. 2021;44:444–57. https://doi.org/10.14348/molcells.2021.0017.
Zhang X, Connelly J, Chao Y, Wang QJ. Multifaceted functions of protein kinase D in pathological processes and human diseases. Biomolecules. 2021;11:483. https://doi.org/10.3390/biom11030483.
Fu Y, Rubin CS. Protein kinase D: coupling extracellular stimuli to the regulation of cell physiology. EMBO Rep. 2011;12:785–96. https://doi.org/10.1038/embor.2011.131.
Storz P, Döppler H, Toker A. Protein kinase D mediates mitochondrion-to-nucleus signaling and detoxification from mitochondrial reactive oxygen species. Mol Cell Biol. 2005;25:8520–30. https://doi.org/10.1128/MCB.25.19.8520-8530.2005.
Eiseler T, Döppler H, Yan IK, Kitatani K, Mizuno K, Storz P. Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat Cell Biol. 2009;11:545–56. https://doi.org/10.1038/ncb1861.
Hausser A, Storz P, Märtens S, Link G, Toker A, Pfizenmaier K. Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIbeta at the Golgi complex. Nat Cell Biol. 2005;7:880–6. https://doi.org/10.1038/ncb1299.
Wakana Y, Campelo F. The PKD-dependent biogenesis of TGN-to-plasma membrane transport carriers. Cells. 2021;10:1618. https://doi.org/10.3390/cells10071618.
Haxhinasto SA, Bishop GA. A novel interaction between protein kinase D and TNF receptor-associated factor molecules regulates B cell receptor-CD40 synergy. J Immunol. 2003;171:4655–62. https://doi.org/10.4049/jimmunol.171.9.4655.
Roy A, Ye J, Deng F, Wang QJ. Protein kinase D signaling in cancer: a friend or foe? Biochim Biophys Acta Rev Cancer. 2017;1868:283–94. https://doi.org/10.1016/j.bbcan.2017.06.004.
Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, et al. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24:8374–85. https://doi.org/10.1128/MCB.24.19.8374-8385.2004.
Fielitz J, Kim MS, Shelton JM, Qi X, Hill JA, Richardson JA, et al. Requirement of protein kinase D1 for pathological cardiac remodeling. Proc Natl Acad Sci USA. 2008;105:3059–63. https://doi.org/10.1073/pnas.0712307105.
Wang S, Li X, Parra M, Verdin E, Bassel-Duby R, Olson EN. Control of endothelial cell proliferation and migration by VEGF signaling to histone deacetylase 7. Proc Natl Acad Sci USA. 2008;105:7738–43. https://doi.org/10.1073/pnas.0803044105.
Xu X, Ha CH, Wong C, Wang W, Hausser A, Pfizenmaier K, et al. Angiotensin II stimulates protein kinase D-dependent histone deacetylase 5 phosphorylation and nuclear export leading to vascular smooth muscle cell hypertrophy. Arterioscler Thromb Vasc Biol. 2007;27:2355–62. https://doi.org/10.1161/ATVBAHA.107.152736.
Jensen ED, Gopalakrishnan R, Westendorf JJ. Bone morphogenic protein 2 activates protein kinase D to regulate histone deacetylase 7 localization and repression of Runx2. J Biol Chem. 2009;284:2225–34. https://doi.org/10.1074/jbc.M805630200.
Wang P, Han L, Shen H, Wang P, Lv C, Zhao G, et al. Protein kinase D1 is essential for Ras-induced senescence and tumor suppression by regulating senescence-associated inflammation. Proc Natl Acad Sci USA. 2014;111:7683–8. https://doi.org/10.1073/pnas.1324267111.
Su Y, Wang P, Shen H, Sun Z, Xu C, Li G, et al. The protein kinase D1-mediated classical protein secretory pathway regulates the Ras oncogene-induced senescence response. J Cell Sci. 2018;131:jcs207217. https://doi.org/10.1242/jcs.207217.
Badreldin AA, Bagheri L, Zhang B, Larson AN, van Wijnen AJ. Relative mRNA and protein stability of epigenetic regulators in musculoskeletal cell culture models. Gene. 2021;766:145032. https://doi.org/10.1016/j.gene.2020.145032.
van Wijnen AJ, Bagheri L, Badreldin AA, Larson AN, Dudakovic A, Thaler R, et al. Biological functions of chromobox (CBX) proteins in stem cell self-renewal, lineage-commitment, cancer and development. Bone. 2021;143:115659. https://doi.org/10.1016/j.bone.2020.115659.
Vandamme J, Völkel P, Rosnoblet C, Le Faou P, Angrand PO. Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells. Mol Cell Proteomics. 2011;10:M110.002642. https://doi.org/10.1074/mcp.M110.002642.
Tamburri S, Lavarone E, Fernández-Pérez D, Conway E, Zanotti M, Manganaro D, et al. Histone H2AK119 mono-ubiquitination is essential for polycomb-mediated transcriptional repression. Mol Cell. 2020;77:840–56. https://doi.org/10.1016/j.molcel.2019.11.021.
Blackledge NP, Fursova NA, Kelley JR, Huseyin MK, Feldmann A, Klose RJ. PRC1 catalytic activity is central to polycomb system function. Mol Cell. 2020;77:857–74. https://doi.org/10.1016/j.molcel.2019.12.006.
Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LLP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–59. https://doi.org/10.1016/j.cell.2014.05.004.
Rose NR, King HW, Blackledge NP, Fursova NA, Ember KJ, Fischer R, et al. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. eLife. 2016;5:e18591. https://doi.org/10.7554/eLife.18591.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31871382 and 32070733) and from the Ministry of Science and Technology of the People’s Republic of China (2018YFC2000102).
Author information
Authors and Affiliations
Contributions
JC conceived the study. JC, ZQF, YYS, and DDL designed the experiments. ZQF performed most of the experiments; YYS conducted Figs. 1A–C, S1, 2C, S2H and S7A, B, D; DDL performed Fig. 1D, F, SILAC experiment, and Fig. 2E, F; FXH performed RNA-seq experiment; RDQ conducted AlphaFold3 prediction. GDL provided resources. ZQF analyzed most of the data and prepared figures. JC, ZQF, and YYS wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Fang, Z., Liu, D., Su, Y. et al. Phosphorylation of CBX8 by PKD1 suppresses PRC1 activity and promotes cell senescence. Oncogene 45, 398–413 (2026). https://doi.org/10.1038/s41388-025-03657-w
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03657-w