Abstract
Background
Prostate cancer (PCa) shows racial disparity in clinical and genomic characteristics, and Asian patients with PCa often present with more aggressive phenotypes at diagnosis. The ability of TP53 to serve as a prognostic biomarker of PCa has been well studied in Western populations. However, no studies to date have examined the role of TP53 in the disparities of primary hormone-naïve prostate cancer (HNPC) between Chinese and Western populations.
Methods
We collected prostate tumors and matched normal tissues or blood samples to perform targeted next-generation sequencing of 94 Chinese primary localized HNPC samples, and correlated these genomic profiles with clinical outcomes. The OncoKB knowledge database was used to identify and classify actionable alterations.
Results
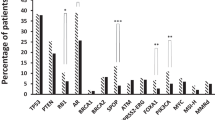
The aberrations of PTEN, CDK12, and SPOP in Chinese HNPC samples were similar to those in the Western samples. However, we demonstrated an association of a high frequency of TP53 alterations (21/94) with a relatively higher percentage of alterations in the Wnt signaling pathway (15/94) in Chinese HNPC. Additionally, we highlighted alterations of LRP1B as accounting for a high proportion of PCa and found more frequent alterations in CDH1 in Chinese PCa. Of these, only CDH1 alteration was associated with rapid biochemical recurrence (BCR). However, we verified that TP53 status was at the core of the genomic alteration landscape in Chinese HNPC with putative driver mutations because of the strong connections with other signaling pathways. The mutually exclusive relationship between alterations in TP53 and Wnt/CTNNB1 further molecularly characterizes subsets of prostate cancers. Moreover, the alteration of KMT2C was more likely to co-occur with TP53 alteration, indicating a more aggressive phenotype of PCa, which was associated with sensitivity to treatment with poly ADT–ribose polymerase (PARP) inhibitors.
Conclusions
Detection of TP53 alterations has clinical utility for guiding precision cancer therapy for HNPC, especially in the Chinese population.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rebecca LS, Kimberly DM, Ahmedin J. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Buttigliero C, Tucci M, Bertaglia V, Vignani F, Bironzo P, Di Maio M, et al. Understanding and overcoming the mechanisms of primary and acquired resistance to abiraterone and enzalutamide in castration resistant prostate cancer. Cancer Treat Rev. 2015;41:884–92.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32.
Chen R, Ren S, Yiu MK, Fai NC, Cheng WS, Ian LH. Chinese Prostate Cancer Consortium et al. Prostate cancer in Asia: a collaborative report. Asian J Urol. 2014;1:15–29.
Ren S, Wei GH, Liu D, Wang L, Hou Y, Zhu S, et al. Whole-genome and transcriptome sequencing of prostate cancer identify new genetic alterations driving disease progression. Eur Urol. 2018;73:322–39.
Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N. Engl J Med. 2014;371:424–33.
de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl J Med. 2011;364:1995–2005.
Oudard S, Fizazi K, Sengeløv L, Daugaard G, Saad F, Hansen S, et al. Cabazitaxel versus docetaxel as first-line therapy for patients with metastatic castration-resistant prostate cancer: a randomized phase III trial-FIRSTANA. J Clin Oncol. 2017;35:3189–97.
Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–17.
Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28.
Hamid AA, Gray KP, Shaw G, MacConaill LE, Evan C, Bernard B, et al. Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. Eur Urol. 2019;76:89–97.
De Laere B, Oeyen S, Mayrhofer M, Whitington T, van Dam PJ, Van Oyen P, et al. TP53 outperforms other androgen receptor biomarkers to predict abiraterone or enzalutamide outcome in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2019;25:1766–73.
Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature. 2017;541:359–64.
Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol. 2013;63:920–6.
Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–25.
Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805.
Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, et al. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N. Engl J Med. 1995;332:1393–8.
Mateo J, Seed G, Bertan C, Rescigno P, Dolling D, Figueiredo I, et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. J Clin Invest. 2020;130:1743–51.
de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for metastatic castration-resistant prostate cancer. N. Engl J Med. 2020;382:2091–102.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl J Med. 2015;372:2509–20.
Abida W, Cheng ML, Armenia J, Middha S, Autio KA, Vargas HA, et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol. 2019;5:471–8.
Nava Rodrigues D, Rescigno P, Liu D, Yuan W, Carreira S, Lambros MB, et al. Immunogenomic analyses associate immunological alterations with mismatch repair defects in prostate cancer. J Clin Invest. 2018;128:4441–53.
Huang JB, Wu YP, Lin YZ, Cai H, Chen SH, Sun XL, et al. Up-regulation of LIMK1 expression in prostate cancer is correlated with poor pathological features, lymph node metastases and biochemical recurrence. J Cell Mol Med. 2020;24:4698–706.
Gleicher S, Kauffman EC, Kotula L, Bratslavsky G, Vourganti S. Implications of high rates of metastatic prostate cancer in BRCA2 mutation carriers. Prostate. 2016;76:1135–45.
Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22.
Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22:369–78.
Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;1:1–16.
Lalonde E, Ishkanian AS, Sykes J, Fraser M, Ross-Adams H, Erho N, et al. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol. 2014;15:1521–32.
Buyyounouski MK, Pickles T, Kestin LL, Allison R, Williams SG. Validating the interval to biochemical failure for the identification of potentially lethal prostate cancer. J Clin Oncol. 2012;30:1857–63.
Ecke TH, Schlechte HH, Schiemenz K, Sachs MD, Lenk SV, Rudolph BD, et al. TP53 gene mutations in prostate cancer progression. Anticancer Res. 2010;30:1579–86.
Tosoian JJ, Almutairi F, Morais CL, Glavaris S, Hicks J, Sundi D, et al. Prevalence and prognostic significance of PTEN loss in African-American and European-American men undergoing radical prostatectomy. Eur Urol. 2017;71:697–700.
Kim SH, Park WS, Yun SI, Joo J, Joung JY, Seo HK, et al. Overexpression of BRCA1 or BRCA2 in prostatectomy specimens is predictive of biochemical recurrence after radical prostatectomy. Histopathology. 2016;68:673–9.
Ateeq B, Bhatia V, Goel S. Molecular discriminators of racial disparities in prostate cancer. Trends Cancer. 2016;2:116–20.
Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, et al. Integrated analysis of TP53 gene and pathway alterations in the cancer genome atlas. Cell Rep. 2019;28:1370–84.e5.
Kunimasa K, Hirotsu Y, Nakamura H, Tamiya M, Iijima Y, Ishida H, et al. Rapid progressive lung cancers harbouring multiple clonal driver mutations with big bang evolution model. Cancer Genet. 2020;241:51–6.
Yamamoto S, Iwakuma T. Regulators of oncogenic mutant TP53 gain of function. Cancers (Basel). 2018;11:4.
Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–5.
Stopsack KH, Nandakumar S, Wibmer AG, Haywood S, Weg ES, Barnett ES, et al. Oncogenic genomic alterations, clinical phenotypes, and outcomes in metastatic castration-sensitive prostate cancer. Clin Cancer Res. 2020;26:3230–8.
Rajan P, Sudbery IM, Villasevil ME, Mui E, Fleming J, Davis M, et al. Next-generation sequencing of advanced prostate cancer treated with androgen-deprivation therapy. Eur Urol. 2014;66:32–9.
Pinyol R, Sia D, Llovet JM. Immune exclusion-Wnt/CTNNB1 class predicts resistance to immunotherapies in HCC. Clin Cancer Res. 2019;25:2021–3.
Linch M, Goh G, Hiley C, Shanmugabavan Y, McGranahan N, Rowan A, et al. Intratumoural evolutionary landscape of high-risk prostate cancer: the PROGENY study of genomic and immune parameters. Ann Oncol. 2017;28:2472–80.
Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75.
Cao H, Liu X, Chen Y, Yang P, Huang T, Song L, et al. Circulating tumor DNA is capable of monitoring the therapeutic response and resistance in advanced colorectal cancer patients undergoing combined target and chemotherapy. Front Oncol. 2020;10:466.
Chen H, Chong W, Wu Q, Yao Y, Mao M, Wang X. Association of LRP1B mutation with tumor mutation burden and outcomes in melanoma and non-small cell lung cancer patients treated with immune check-point blockades. Front Immunol. 2019;10:1113.
Tucker MD, Zhu J, Marin D, Gupta RT, Gupta S, Berry WR, et al. Pembrolizumab in men with heavily treated metastatic castrate-resistant prostate cancer. Cancer Med. 2019;8:4644–55.
Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8:444–57.
Lindberg J, Mills IG, Klevebring D, Liu W, Neiman M, Xu J, et al. The mitochondrial and autosomal mutation landscapes of prostate cancer. Eur Urol. 2013;63:702–8.
Rampias T, Karagiannis D, Avgeris M, Polyzos A, Kokkalis A, Kanaki Z, et al. The lysine-specific methyltransferase KMT2C/MLL3 regulates DNA repair components in cancer. EMBO Rep. 2019;20:e46821.
Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–68.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (NO.81800672) and the Key Research and Development Program of Shandong Province (NO.2017GSF18105 and NO.2019GSF108123).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Z., Guo, H., Zhu, Y. et al. TP53 alterations of hormone-naïve prostate cancer in the Chinese population. Prostate Cancer Prostatic Dis 24, 482–491 (2021). https://doi.org/10.1038/s41391-020-00302-3
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41391-020-00302-3
This article is cited by
-
Comprehensive genomic landscape of metastatic hormone-sensitive prostate cancer in the Chinese population
World Journal of Surgical Oncology (2025)
-
The relationship between p53 and the malignant transformation of nasal inverted papilloma: a meta-analysis
Egyptian Journal of Medical Human Genetics (2025)
-
A Recent Overview of Molecular Pathways in Synthetic Lethality as a Proposed Valid Target in Oncology: Current Insights and Future Directions
Indian Journal of Surgical Oncology (2025)
-
Prediction of lung adenocarcinoma prognosis and clinical treatment efficacy by telomere-associated gene risk model
Discover Oncology (2025)
-
Network pharmacology and experimental validation to explore the role and potential mechanism of Liuwei Dihuang Decoction in prostate cancer
BMC Complementary Medicine and Therapies (2024)