Abstract
Background
This study aimed to develop and validate deep learning (DL) models based on multiparametric MRI (mpMRI) and [18F]PSMA-1007 PET/CT to predict extracapsular extension (ECE) in prostate cancer (PCa), and to explore easy models integrating DL with clinical expertise.
Methods
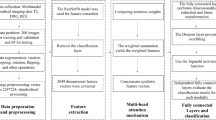
A total of 388 patients who underwent radical prostatectomy were enrolled from centers A, B and C. Three DL models based on mpMRI, PET/CT, and a combined MPC model were developed and compared with a manual model based on the ECE grading system. Additionally, three combined models (mpMRI-M, PET/CT-M, and MPC-M) were constructed by integrating the DL models with the Manual model. To enhance clinical applicability, an easy model (E-MPC-M) was developed. Model performance was evaluated using the area under the receiver-operating-characteristic curve (AUC) and metrics derived from the confusion matrix. Gradient-weighted class-activation-mapping (Grad-CAM) was employed to visualize model interpretability.
Results
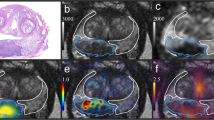
In the internal cohort, the Manual, MPC, and MPC-M models achieved AUCs of 0.752, 0.897, and 0.907, respectively; corresponding sensitivities were 0.616, 0.896, and 0.915, and specificities were 0.791, 0.740, and 0.802. In the external validation cohort, these models achieved AUCs of 0.665, 0.824, and 0.849; sensitivities of 0.318, 0.955, and 0.955; and specificities of 0.960, 0.600, and 0.640, respectively. The E-MPC-M model also showed robust performance, with an AUC of 0.862 in the internal cohort and 0.775 in the external cohort. Grad-CAM visualizations highlighted the model’s focus on tumor-relevant regions, confirming effective learning of tumor features.
Conclusions
The MPC-M model demonstrated strong predictive performance for PCa ECE across internal and external cohorts, while the E-MPC-M model retained much of this performance with enhanced clinical practicality. However, these models should be considered as preliminary, and larger prospective multicenter studies are required to confirm their robustness and generalizability.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
Code availability
The custom Python code used to generate the results and central conclusions of this study is available from the corresponding author upon reasonable request.
References
Siegel RL, Kratzer TB, Giaquinto AN, Sung H, Jemal A. Cancer statistics, 2025. CA Cancer J Clin. 2025;75:10–45.
Sayyid R, Perlis N, Ahmad A, Evans A, Toi A, Horrigan M, et al. Development and external validation of a biopsy-derived nomogram to predict risk of ipsilateral extraprostatic extension. BJU Int. 2017;120:76–82.
Mottet N, van den Bergh RCN, Briers E, Van den Broeck T, Cumberbatch MG, De Santis M, et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur Urol. 2021;79:243–62.
Bill-Axelson A, Holmberg L, Garmo H, Taari K, Busch C, Nordling S, et al. Radical Prostatectomy or Watchful Waiting in Prostate Cancer - 29-Year Follow-up. N Engl J Med. 2018;379:2319–29.
Jeong BC, Chalfin HJ, Lee SB, Feng Z, Epstein JI, Trock BJ, et al. The relationship between the extent of extraprostatic extension and survival following radical prostatectomy. Eur Urol. 2015;67:342–6.
Somford DM, Hamoen EH, Fütterer JJ, van Basten JP, Hulsbergen-van de Kaa CA, Vreuls W, et al. The predictive value of endorectal 3 Tesla multiparametric magnetic resonance imaging for extraprostatic extension in patients with low, intermediate and high risk prostate cancer. J Urol. 2013;190:1728–34.
Walz J, Epstein JI, Ganzer R, Graefen M, Guazzoni G, Kaouk J, et al. A Critical Analysis of the Current Knowledge of Surgical Anatomy of the Prostate Related to Optimisation of Cancer Control and Preservation of Continence and Erection in Candidates for Radical Prostatectomy: An Update. Eur Urol. 2016;70:301–11.
Patel VR, Sandri M, Grasso AAC, De Lorenzis E, Palmisano F, Albo G, et al. A novel tool for predicting extracapsular extension during graded partial nerve sparing in radical prostatectomy. BJU Int. 2018;121:373–82.
Egawa S, Suyama K, Matsumoto K, Satoh T, Uchida T, Kuwao S, et al. Improved predictability of extracapsular extension and seminal vesicle involvement based on clinical and biopsy findings in prostate cancer in Japanese men. Urology. 1998;52:433–40.
Egawa S, Suyama K, Arai Y, Matsumoto K, Tsukayama C, Kuwao S, et al. A study of pretreatment nomograms to predict pathological stage and biochemical recurrence after radical prostatectomy for clinically resectable prostate cancer in Japanese men. Jpn J Clin Oncol. 2001;31:74–81.
Song C, Kang T, Ro JY, Lee MS, Kim CS, Ahn H. Nomograms for the prediction of pathologic stage of clinically localized prostate cancer in Korean men. J Korean Med Sci. 2005;20:262–6.
Huang Y, Isharwal S, Haese A, Chun FK, Makarov DV, Feng Z, et al. Prediction of patient-specific risk and percentile cohort risk of pathological stage outcome using continuous prostate-specific antigen measurement, clinical stage and biopsy Gleason score. BJU Int. 2011;107:1562–9.
Satake N, Ohori M, Yu C, Kattan MW, Ohno Y, Miyakawa A, et al. Development and internal validation of a nomogram predicting extracapsular extension in radical prostatectomy specimens. Int J Urol. 2010;17:267–72.
Eissa A, Elsherbiny A, Zoeir A, Sandri M, Pirola G, Puliatti S, et al. Reliability of the different versions of Partin tables in predicting extraprostatic extension of prostate cancer: a systematic review and meta-analysis. Minerva Urologica e Nefrologica = Ital J Urol Nephrol. 2019;71:457–78.
Steuber T, Graefen M, Haese A, Erbersdobler A, Chun FK, Schlom T, et al. Validation of a nomogram for prediction of side specific extracapsular extension at radical prostatectomy. J Urol. 2006;175:939–44.
Zanelli E, Giannarini G, Cereser L, Zuiani C, Como G, Pizzolitto S, et al. Head-to-head comparison between multiparametric MRI, the partin tables, memorial sloan kettering cancer center nomogram, and CAPRA score in predicting extraprostatic cancer in patients undergoing radical prostatectomy. J Magn Reson Imaging. 2019;50:1604–13.
Ohori M, Kattan MW, Koh H, Maru N, Slawin KM, Shariat S, et al. Predicting the presence and side of extracapsular extension: a nomogram for staging prostate cancer. J Urol. 2004;171:1844–9.
Rocco B, Sighinolfi MC, Sandri M, Eissa A, Elsherbiny A, Zoeir A, et al. Is Extraprostatic Extension of Cancer Predictable? A Review of Predictive Tools and an External Validation Based on a Large and a Single Center Cohort of Prostate Cancer Patients. Urology. 2019;129:8–20.
Morlacco A, Sharma V, Viers BR, Rangel LJ, Carlson RE, Froemming AT, et al. The Incremental Role of Magnetic Resonance Imaging for Prostate Cancer Staging before Radical Prostatectomy. Eur Urol. 2017;71:701–4.
Fang D, Zhao C, Ren D, Yu W, Wang R, Wang H, et al. Could Magnetic Resonance Imaging Help to Identify the Presence of Prostate Cancer Before Initial Biopsy? The Development of Nomogram Predicting the Outcomes of Prostate Biopsy in the Chinese Population. Ann Surg Oncol. 2016;23:4284–92.
Foley RW, Redman SL, Graham RN, Loughborough WW, Little D. Fluorine-18 labelled prostate-specific membrane antigen (PSMA)-1007 positron-emission tomography-computed tomography: normal patterns, pearls, and pitfalls. Clin Radio. 2020;75:903–13.
Awenat S, Piccardo A, Carvoeiras P, Signore G, Giovanella L, Prior JO, et al. Diagnostic Role of (18)F-PSMA-1007 PET/CT in prostate cancer staging: a systematic review. Diagnostics 2021;11:552.
Huang S, Ong S, McKenzie D, Mirabelli A, Chen DC, Chengodu T, et al. Comparison of 18F-based PSMA radiotracers with [68Ga]Ga-PSMA-11 in PET/CT imaging of prostate cancer—a systematic review and meta-analysis. Prostate Cancer Prostatic Dis. 2023;27:654–64.
Chan HP, Samala RK, Hadjiiski LM, Zhou C. Deep Learning in Medical Image Analysis. Adv Exp Med Biol. 2020;1213:3–21.
Hesamian MH, Jia W, He X, Kennedy P. Deep learning techniques for medical image segmentation: achievements and challenges. J Digital Imaging. 2019;32:582–96.
Schelb P, Kohl S, Radtke JP, Wiesenfarth M, Kickingereder P, Bickelhaupt S, et al. Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology. 2019;293:607–17.
Moroianu ŞL, Bhattacharya I, Seetharaman A, Shao W, Kunder CA, Sharma A, et al. Computational detection of extraprostatic extension of prostate cancer on multiparametric MRI using deep learning. Cancers 2022;14:2821.
Baydoun A, Jia AY, Zaorsky NG, Kashani R, Rao S, Shoag JE, et al. Artificial intelligence applications in prostate cancer. Prostate Cancer Prostatic Dis. 2024;27:37–45.
Mehralivand S, Shih JH, Harmon S, Smith C, Bloom J, Czarniecki M, et al. A Grading System for the Assessment of Risk of Extraprostatic Extension of Prostate Cancer at Multiparametric MRI. Radiology. 2019;290:709–19.
Ben-David S, Blitzer J, Crammer K, Kulesza A, Pereira F. Vaughan JWJML. A Theory Learn Differ Domains. 2010;79:151–75.
Castro DC, Walker I, Glocker B. Causality matters in medical imaging. Nat Commun. 2020;11:3673.
Unsupervised Domain Adaptation in brain lesion segmentation with adversarial networks. International Conference on Information Processing in Medical Imaging 2017.
Mehta P, Antonelli M, Ahmed HU, Emberton M, Analysis SOJMI. Computer-aided diagnosis of prostate cancer using multiparametric MRI and clinical features: A patient-level classification framework. Med Image Anal. 2021;73:102153.
Zhao L, Bao J, Qiao X, Jin P, Ji Y, Li Z, et al. Predicting clinically significant prostate cancer with a deep learning approach: a multicentre retrospective study. Eur J Nucl Med Mol Imaging. 2023;50:727–41.
He T, Guo J, Chen N, Xu X, Wang Z, Fu K, et al. MediMLP: Using Grad-CAM to Extract crucial variables for lung cancer postoperative complication. Prediction. 2020;2410.
Rodriguez-Sanchez L, Martini A, Zhuang J, Guo H, Rajwa P, Mandoorah Q, et al. External validation of an algorithm to personalize nerve sparing approaches during robot-assisted radical prostatectomy in men with unilateral high-risk prostate cancer. Prostate Cancer Prostatic Dis. 2025;28:223–5.
Tillu N, Maheshwari A, Kolanukuduru K, Choudhary M, Agarwal Y, Joshi H, et al. Predicting side-specific extraprostatic extension in prostate cancer using an 18F-DCFPyL PSMA-PET/CT-based nomogram. Prostate Cancer Prostatic Dis. 2025. https://doi.org/10.1038/s41391-025-01001-7.
Prata F, Anceschi U, Cordelli E, Faiella E, Civitella A, Tuzzolo P, et al. Radiomic Machine-Learning Analysis of Multiparametric Magnetic Resonance Imaging in the Diagnosis of Clinically Significant Prostate Cancer: New Combination of Textural and Clinical Features. Curr Oncol. 2023;30:2021–31.
Santucci D, Ragone R, Vergantino E, Vaccarino F, Esperto F, Prata F, et al. Comparison between Three Radiomics Models and Clinical Nomograms for Prediction of Lymph Node Involvement in PCa Patients Combining Clinical and Radiomic Features. Cancers 2024;16:2731.
Li EV, Schaeffer EM, Ramesh Kumar SKS, Zhou R, Yang XJ, Mana-Ay M, et al. Utility of 18F-DCFPyL PET for local staging for high or very high risk prostate cancer for patients undergoing radical prostatectomy. Eur J Nucl Med Mol Imaging. 2025;52:2335–42.
Pan K, Yao F, Hong W, Xiao J, Bian S, Zhu D, et al. Multimodal radiomics based on 18F-Prostate-specific membrane antigen-1007 PET/CT and multiparametric MRI for prostate cancer extracapsular extension prediction. Br J Radiol 2024;97:408–14.
Yao F, Lin H, Xue Y-N, Zhuang Y-D, Bian S-Y, Zhang Y-Y, et al. Multimodal imaging deep learning model for predicting extraprostatic extension in prostate cancer using MpMRI and 18 F-PSMA-PET/CT. Cancer Imaging 2025;25:103.
Sung J. Artificial intelligence in medicine: Ethical, social and legal perspectives. Ann Acad Med Singap. 2023;52:695–9.
Naik N, Hameed BMZ, Shetty DK, Swain D, Shah M, Paul R, et al. Legal and Ethical Consideration in Artificial Intelligence in Healthcare: Who Takes Responsibility?. Front Surg. 2022;9:862322.
Sallam M. ChatGPT Utility in Healthcare Education, research, and practice: systematic review on the promising perspectives and valid concerns. Healthcare 2023;11:887.
Bhargava DC, Jadav D, Meshram VP, Kanchan T. ChatGPT in medical research: challenging time ahead. Med-Leg J. 2023;91:223–5.
Funding
This study has received funding by the Key Laboratory of Novel Nuclide Technologies on Precision Diagnosis and Treatment & clinical Transformation of Wenzhou (Grant No. 2023HZSY0012), the Discipline Cluster of Oncology, Wenzhou Medical University, China (Grant No. z1-2023008), the Summit Advancement Disciplines of Zhejiang Province (Wenzhou Medical University – Pharmaceutics).
Author information
Authors and Affiliations
Contributions
Conception and design: FY and KHP. Acquisition of data: QL, TCL, SYB, and YDZ. Analysis and interpretation of data: DQZ, HL, and CKM. Drafting of the manuscript: YF and DQZ. Critical revision of the manuscript: DQZ and KHP. Statistical analysis: YF, HL, and CKM. Supervision: YYJ and JL.
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Ethics approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations. This study was approved by our hospital’s Institutional Review Board (Reference Number: KY2022-R012). Since this study was a retrospective study, the requirement for informed consent was waived.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yao, F., Zhu, D., Lin, H. et al. Deep learning algorithm assisting diagnosis of prostate cancer extracapsular extension based on [18F]PSMA-1007 PET/CT and multiparametric MRI: A multicenter study. Prostate Cancer Prostatic Dis (2025). https://doi.org/10.1038/s41391-025-01063-7
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41391-025-01063-7