Abstract
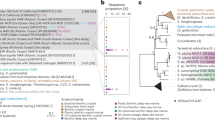
Deep-sea hydrothermal plumes are considered natural laboratories for understanding ecological and biogeochemical interactions. Previous studies focused on interactions between microorganisms and inorganic, reduced hydrothermal inputs including sulfur, hydrogen, iron, and manganese. However, little is known about transformations of organic compounds, especially methylated, sulfur-containing compounds, and petroleum hydrocarbons. Here, we reconstructed nine gammaproteobacterial metagenome-assembled genomes, affiliated with Methylococcales, Methylophaga, and Cycloclasticus, from three hydrothermal ecosystems. We present evidence that these three groups have high transcriptional activities of genes encoding cycling of C1-compounds, petroleum hydrocarbons, and organic sulfur in hydrothermal plumes. This includes oxidation of methanethiol, the simplest thermochemically-derived organic sulfur, for energy metabolism in Methylococcales and Cycloclasticus. Together with active transcription of genes for thiosulfate and methane oxidation in Methylococcales, these results suggest an adaptive strategy of versatile and simultaneous use of multiple available electron donors. Meanwhile, the first near-complete MAG of hydrothermal Methylophaga aminisulfidivorans and its transcriptional profile point to active chemotaxis targeting small organic compounds. Petroleum hydrocarbon-degrading Cycloclasticus are abundant and active in plumes of oil spills as well as deep-sea vents, suggesting that they are indigenous and effectively respond to stimulus of hydrocarbons in the deep sea. These findings suggest that these three groups of Gammaproteobacteria transform organic carbon and sulfur compounds via versatile and opportunistic metabolism and modulate biogeochemistry in plumes of hydrothermal systems as well as oil spills, thus contributing broad ecological impact to the deep ocean globally.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
The Sequence Read Archive IDs for metagenomic and metatranscriptomic are summarized in Supplementary Dataset S3. The MAGs that were resolved from this study are deposited under NCBI BioProject PRJNA488180. The further analyses on gene operons of methyl-metabolism, transcriptomic ratio of Hydro-γ-MAGs, and Cycloclasticus expression in DWH are present in Supplementary Information and Figs. S11–S18.
References
Dick GJ. The microbiomes of deep-sea hydrothermal vents: distributed globally, shaped locally. Nat Rev Microbiol. 2019;17:271–83.
Reeves EP, McDermott JM, Seewald JS. The origin of methanethiol in midocean ridge hydrothermal fluids. Proc Natl Acad Sci USA. 2014;111:5474.
Lang SQ, Butterfield DA, Schulte M, Kelley DS, Lilley MD. Elevated concentrations of formate, acetate and dissolved organic carbon found at the Lost City hydrothermal field. Geochim Cosmochim Acta. 2010;74:941–52.
Konn C, Charlou JL, Donval JP, Holm NG, Dehairs F, Bouillon S. Hydrocarbons and oxidized organic compounds in hydrothermal fluids from Rainbow and Lost City ultramafic-hosted vents. Chem Geol. 2009;258:299–314.
Proskurowski G, Lilley MD, Seewald JS, Fruh-Green GL, Olson EJ, Lupton JE, et al. Abiogenic hydrocarbon production at Lost City hydrothermal field. Science. 2008;319:604–7.
Dick G, Anantharaman K, Baker B, Li M, Reed D, Sheik C. The microbiology of deep-sea hydrothermal vent plumes: ecological and biogeographic linkages to seafloor and water column habitats. Front Microbiol. 2013;4:124.
Baker BJ, Lazar CS, Teske AP, Dick GJ. Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome. 2015;3:14.
Dombrowski N, Seitz KW, Teske AP, Baker BJ. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome. 2017;5:106.
Dombrowski N, Teske AP, Baker BJ. Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat Commun. 2018;9:4999.
Zhou Z, Liu Y, Xu W, Pan J, Luo Z-H, Li M. Genome and community-level interaction insights on wide carbon utilizing and element cycling function of Hydrothermarchaeota from hydrothermal sediment. mSystems. 2020;5:e00795–00719.
Baker BJ, Lesniewski RA, Dick GJ. Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume. ISME J. 2012;6:2269.
Anantharaman K, Breier JA, Sheik CS, Dick GJ. Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc Natl Acad Sci USA. 2013;110:330.
Lesniewski RA, Jain S, Anantharaman K, Schloss PD, Dick GJ. The metatranscriptome of a deep-sea hydrothermal plume is dominated by water column methanotrophs and lithotrophs. ISME J. 2012;6:2257.
Anantharaman K, Duhaime MB, Breier JA, Wendt K, Toner BM, Dick GJ. Sulfur oxidation genes in diverse deep-sea viruses. Science. 2014;344:757–60.
Li M, Jain S, Dick GJ. Genomic and transcriptomic resolution of organic matter utilization among deep-sea bacteria in guaymas basin hydrothermal plumes. Front Microbiol. 2016;7:1125.
Li M, Baker BJ, Anantharaman K, Jain S, Breier JA, Dick GJ. Genomic and transcriptomic evidence for scavenging of diverse organic compounds by widespread deep-sea archaea. Nat Commun. 2015;6:8933.
Li M, Jain S, Baker BJ, Taylor C, Dick GJ. Novel hydrocarbon monooxygenase genes in the metatranscriptome of a natural deep-sea hydrocarbon plume. Environ Microbiol. 2014;16:60–71.
Skennerton CT, Ward LM, Michel A, Metcalfe K, Valiente C, Mullin S, et al. Genomic reconstruction of an uncultured hydrothermal vent gammaproteobacterial methanotroph (Family Methylothermaceae) indicates multiple adaptations to oxygen limitation. Front Microbiol. 2015;6:1425.
Villeneuve C, Martineau C, Mauffrey F, Villemur R. Methylophaga nitratireducenticrescens sp. nov. and Methylophaga frappieri sp. nov., isolated from the biofilm of the methanol-fed denitrification system treating the seawater at the Montreal Biodome. Int J Syst Evol Microbiol. 2013;63:2216–22.
Rivers AR, Sharma S, Tringe SG, Martin J, Joye SB, Moran MA. Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J. 2013;7:2315.
Sargeant S, Murrell J, Nightingale P, Dixon J. Seasonal variability in microbial methanol utilisation in coastal waters of the western English Channel. Mar Ecol Prog Ser. 2016;550:53–64.
Ruff SE, Biddle JF, Teske AP, Knittel K, Boetius A, Ramette A. Global dispersion and local diversification of the methane seep microbiome. Proc Natl Acad Sci USA. 2015;112:4015–20.
Rubin-Blum M, Antony CP, Borowski C, Sayavedra L, Pape T, Sahling H, et al. Short-chain alkanes fuel mussel and sponge Cycloclasticus symbionts from deep-sea gas and oil seeps. Nat Microbiol. 2017;2:17093.
Schneiker S, Martins dos Santos VA, Bartels D, Bekel T, Brecht M, Buhrmester J, et al. Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat Biotechnol. 2006;24:997–1004.
Lai Q, Li W, Shao Z. Complete genome sequence of Alcanivorax dieselolei type strain B5. J Bacteriol. 2012;194:6674.
Rani S, Jeon WJ, Koh HW, Kim YE, Kang MS, Park SJ. Genomic potential of Marinobacter salinus Hb8(T) as sulfur oxidizing and aromatic hydrocarbon degrading bacterium. Mar Genom. 2017;34:19–21.
Singer E, Webb EA, Nelson WC, Heidelberg JF, Ivanova N, Pati A, et al. Genomic potential of marinobacter aquaeolei, a biogeochemical “Opportunitroph”. Appl Environ Microbiol. 2011;77:2763–71.
Fondi M, Rizzi E, Emiliani G, Orlandini V, Berna L, Papaleo MC, et al. The genome sequence of the hydrocarbon-degrading Acinetobacter venetianus VE-C3. Res Microbiol. 2013;164:439–49.
Jung J, Baek JH, Park W. Complete genome sequence of the diesel-degrading Acinetobacter sp. strain DR1. J Bacteriol. 2010;192:4794–5.
Sheik CS, Jain S, Dick GJ. Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ Microbiol. 2013;16:304–17.
Breier JA, Sheik CS, Gomez-Ibanez D, Sayre-McCord RT, Sanger R, Rauch C, et al. A large volume particulate and water multi-sampler with in situ preservation for microbial and biogeochemical studies. Deep Sea Res Part I: Oceanographic Res Pap. 2014;94:195–206.
Breier JA, Rauch CG, McCartney K, Toner BM, Fakra SC, White SN, et al. A suspended-particle rosette multi-sampler for discrete biogeochemical sampling in low-particle-density waters. Deep Sea Res Part I. 2009;56:1579–89.
Breier J, Toner B, Fakra S, Marcus M, White S, Thurnherr A, et al. Sulfur, sulfides, oxides and organic matter aggregated in submarine hydrothermal plumes at 9°50′N East Pacific Rise. Geochim Cosmochim Acta. 2012;88:216–36.
Dick GJ, Tebo BM. Microbial diversity and biogeochemistry of the Guaymas Basin deep-sea hydrothermal plume. Environ Microbiol. 2010;12:1334–47.
Sheik CS, Anantharaman K, Breier JA, Sylvan JB, Edwards KJ, Dick GJ. Spatially resolved sampling reveals dynamic microbial communities in rising hydrothermal plumes across a back-arc basin. ISME J. 2014;9:1434.
Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357.
Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165.
Sieber CM, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol. 2018;3:836–43.
Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–42.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55.
Laczny CC, Sternal T, Plugaru V, Gawron P, Atashpendar A, Margossian HH, et al. VizBin-an application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome, 2015;3:1.
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185.
Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–31.
Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–D293.
Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60.
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2014;32:268–74.
Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403.
Teske A, de Beer D, McKay LJ, Tivey MK, Biddle JF, Hoer D, et al. The guaymas basin hiking guide to hydrothermal mounds, chimneys, and microbial mats: complex seafloor expressions of subsurface hydrothermal circulation. Front Microbiol. 2016;7:75.
Reveillaud J, Reddington E, McDermott J, Algar C, Meyer JL, Sylva S, et al. Subseafloor microbial communities in hydrogen-rich vent fluids from hydrothermal systems along the Mid-Cayman Rise. Environ Microbiol. 2016;18:1970–87.
Anantharaman K, Breier JA, Dick GJ. Metagenomic resolution of microbial functions in deep-sea hydrothermal plumes across the Eastern Lau Spreading Center. ISME J. 2015;10:225.
Eyice O, Myronova N, Pol A, Carrion O, Todd JD, Smith TJ, et al. Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere. ISME J. 2018;12:145–60.
Wang Y, Graichen ME, Liu A, Pearson AR, Wilmot CM, Davidson VL. MauG, a novel diheme protein required for tryptophan tryptophylquinone biogenesis. Biochemistry. 2003;42:7318–25.
Gondwe M, Krol M, Klaassen W, Gieskes W, de Baar H. Comparison of modeled versus measured MSA:nss SO4 = ratios: a global analysis. Global Biogeochem Cycles 2004;18:2.
Li M, Toner BM, Baker BJ, Breier JA, Sheik CS, Dick GJ. Microbial iron uptake as a mechanism for dispersing iron from deep-sea hydrothermal vents. Nat Commun. 2014;5:3192.
Mason OU, Hazen TC, Borglin S, Chain PS, Dubinsky EA, Fortney JL, et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 2012;6:1715–27.
Yooseph S, Nealson KH, Rusch DB, McCrow JP, Dupont CL, Kim M, et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468:60.
Haro-Moreno JM, Rodriguez-Valera F, Lopez-Garcia P, Moreira D, Martin-Cuadrado A-B. New insights into marine group III Euryarchaeota, from dark to light. ISME J. 2017;11:1102–17.
Larsson J, Celepli N, Ininbergs K, Dupont CL, Yooseph S, Bergman B, et al. Picocyanobacteria containing a novel pigment gene cluster dominate the brackish water Baltic Sea. ISME J. 2014;8:1892.
Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–8.
Keltjens JT, Pol A, Reimann J, Op den Camp HJM. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol. 2014;98:6163–83.
Carrión O, Curson ARJ, Kumaresan D, Fu Y, Lang AS, Mercadé E, et al. A novel pathway producing dimethylsulphide in bacteria is widespread in soil environments. Nat Commun. 2015;6:6579.
Cron BR, Sheik CS, Kafantaris FCA, Druschel GK, Seewald JS, German CR, et al. Dynamic biogeochemistry of the particulate sulfur pool in a buoyant deep-sea hydrothermal plume. ACS Earth Space Chem. 2020;4:168–82.
Meier DV, Pjevac P, Bach W, Hourdez S, Girguis PR, Vidoudez C, et al. Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J. 2017;11:1545.
Chen X-G, Lyu S-S, Garbe-Schönberg D, Lebrato M, Li X, Zhang H-Y, et al. Heavy metals from Kueishantao shallow-sea hydrothermal vents, offshore northeast Taiwan. J Mar Syst. 2016;180:211–9.
McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA. 2010;107:16420–7.
Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7:13219.
Hansell DA, Carlson CA, Repeta DJ, Schlitzer R. Dissolved organic matter in the ocean a controversy stimulates new insights. Oceanography. 2009;22:202–11.
Arrigo KR. Marine microorganisms and global nutrient cycles. Nature. 2004;437:349–55.
Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–8.
Chen Y, Patel NA, Crombie A, Scrivens JH, Murrell JC. Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc Natl Acad Sci USA. 2011;108:17791–6.
Leahy JG, Batchelor PJ, Morcomb SM. Evolution of the soluble diiron monooxygenases. FEMS Microbiol Rev. 2003;27:449–79.
Acknowledgements
Thanks to the suggestions and critical reviews from anonymous reviewers. Thanks to the proofreading efforts and comments from Patricia Tran from University of Wisconsin – Madison. This study was funded by National Natural Science Foundation of China (No. 91851105, 31970105), the Science and Technology Innovation Committee of Shenzhen (No. JCYJ20170818091727570), the Key Project of Department of Education of Guangdong Province (No. 2017KZDXM071), Gordon and Betty Moore Foundation (GBMF2609 to GJD), and the National Science Foundation (OCE 1029242). We are grateful to the captains, crew, and chief scientists (Fred Prahl, Brian Popp, Anna-Louise Reysenbach, and Chris German) of the cruises on which samples were collected.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhou, Z., Liu, Y., Pan, J. et al. Gammaproteobacteria mediating utilization of methyl-, sulfur- and petroleum organic compounds in deep ocean hydrothermal plumes. ISME J 14, 3136–3148 (2020). https://doi.org/10.1038/s41396-020-00745-5
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-020-00745-5
This article is cited by
-
Diverse quorum sensing systems regulate microbial communication and biogeochemical processes in deep-sea cold seeps
Microbiome (2025)
-
Macroalgal ecosystem provides a scalable solution to coastal herbicide pollution via macroalga–microbiome synergy
Communications Earth & Environment (2025)
-
Elevated heterotrophic activity in Guaymas Basin hydrothermal plumes influences deep-sea carbon cycling
Nature Communications (2025)
-
Evaluation of microbial diversity in the formation water of the producer and marginal wells in bokaro coal field
Scientific Reports (2024)
-
Widespread and active piezotolerant microorganisms mediate phenolic compound degradation under high hydrostatic pressure in hadal trenches
Marine Life Science & Technology (2024)