Abstract
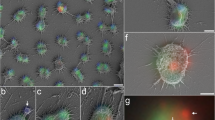
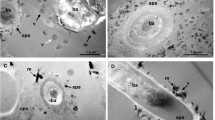
Viruses of hyperthermophilic archaea represent one of the least understood parts of the virosphere, showing little genomic and morphological similarity to viruses of bacteria or eukaryotes. Here, we investigated virus diversity in the active sulfurous fields of the Campi Flegrei volcano in Pozzuoli, Italy. Virus-like particles displaying eight different morphotypes, including lemon-shaped, droplet-shaped and bottle-shaped virions, were observed and five new archaeal viruses proposed to belong to families Rudiviridae, Globuloviridae and Tristromaviridae were isolated and characterized. Two of these viruses infect neutrophilic hyperthermophiles of the genus Pyrobaculum, whereas the remaining three have rod-shaped virions typical of the family Rudiviridae and infect acidophilic hyperthermophiles belonging to three different genera of the order Sulfolobales, namely, Saccharolobus, Acidianus, and Metallosphaera. Notably, Metallosphaera rod-shaped virus 1 is the first rudivirus isolated on Metallosphaera species. Phylogenomic analysis of the newly isolated and previously sequenced rudiviruses revealed a clear biogeographic pattern, with all Italian rudiviruses forming a monophyletic clade, suggesting geographical structuring of virus communities in extreme geothermal environments. Analysis of the CRISPR spacers suggests that isolated rudiviruses have experienced recent host switching across the genus boundary, potentially to escape the targeting by CRISPR-Cas immunity systems. Finally, we propose a revised classification of the Rudiviridae family, with the establishment of six new genera. Collectively, our results further show that high-temperature continental hydrothermal systems harbor a highly diverse virome and shed light on the evolution of archaeal viruses.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
Genome sequences of the isolated viruses have been deposited in GenBank and their accession numbers are listed in Table 2.
References
Prangishvili D, Bamford DH, Forterre P, Iranzo J, Koonin EV, Krupovic M. The enigmatic archaeal virosphere. Nat Rev Microbiol. 2017;15:724–39.
Munson-McGee JH, Snyder JC, Young MJ. Archaeal viruses from high-temperature environments. Genes (Basel). 2018;9:E128.
Wang H, Peng N, Shah SA, Huang L, She Q. Archaeal extrachromosomal genetic elements. Microbiol Mol Biol Rev. 2015;79:117–52.
Dellas N, Snyder JC, Bolduc B, Young MJ. Archaeal Viruses: Diversity, Replication, and Structure. Annu Rev Virol. 2014;1:399–426.
Wang H, Guo Z, Feng H, Chen Y, Chen X, Li Z, et al. Novel Sulfolobus virus with an exceptional capsid architecture. J Virol. 2018;92:e01727–17.
Krupovic M, Quemin ER, Bamford DH, Forterre P, Prangishvili D. Unification of the globally distributed spindle-shaped viruses of the Archaea. J Virol. 2014;88:2354–8.
Contursi P, Fusco S, Cannio R, She Q. Molecular biology of fuselloviruses and their satellites. Extremophiles. 2014;18:473–89.
Bath C, Dyall-Smith ML. His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J Virol. 1998;72:9392–5.
Gorlas A, Koonin EV, Bienvenu N, Prieur D, Geslin C. TPV1, the first virus isolated from the hyperthermophilic genus Thermococcus. Environ Microbiol. 2012;14:503–16.
Geslin C, Le Romancer M, Erauso G, Gaillard M, Perrot G, Prieur D. PAV1, the first virus-like particle isolated from a hyperthermophilic euryarchaeote, “Pyrococcus abyssi”. J Bacteriol. 2003;185:3888–94.
Kim JG, Kim SJ, Cvirkaite-Krupovic V, Yu WJ, Gwak JH, Lopez-Perez M, et al. Spindle-shaped viruses infect marine ammonia-oxidizing thaumarchaea. Proc Natl Acad Sci USA. 2019;116:15645–50.
Rice G, Stedman K, Snyder J, Wiedenheft B, Willits D, Brumfield S, et al. Viruses from extreme thermal environments. Proc Natl Acad Sci USA. 2001;98:13341–5.
Liu Y, Ishino S, Ishino Y, Pehau-Arnaudet G, Krupovic M, Prangishvili D. A novel type of polyhedral viruses infecting hyperthermophilic archaea. J Virol. 2017;91:e00589–17.
Wagner C, Reddy V, Asturias F, Khoshouei M, Johnson JE, Manrique P, et al. Isolation and characterization of Metallosphaera turreted icosahedral virus, a founding member of a new family of archaeal viruses. J Virol. 2017;91:e00925–17.
Wang F, Liu Y, Su Z, Osinski T, de Oliveira GAP, Conway JF, et al. A packing for A-form DNA in an icosahedral virus. Proc Natl Acad Sci USA. 2019;116:22591–7.
Maaty WS, Ortmann AC, Dlakic M, Schulstad K, Hilmer JK, Liepold L, et al. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J Virol. 2006;80:7625–35.
Liu Y, Osinski T, Wang F, Krupovic M, Schouten S, Kasson P, et al. Structural conservation in a membrane-enveloped filamentous virus infecting a hyperthermophilic acidophile. Nat Commun. 2018;9:3360.
Hochstein R, Bollschweiler D, Dharmavaram S, Lintner NG, Plitzko JM, Bruinsma R, et al. Structural studies of Acidianus tailed spindle virus reveal a structural paradigm used in the assembly of spindle-shaped viruses. Proc Natl Acad Sci USA. 2018;115:2120–5.
Ptchelkine D, Gillum A, Mochizuki T, Lucas-Staat S, Liu Y, Krupovic M, et al. Unique architecture of thermophilic archaeal virus APBV1 and its genome packaging. Nat Commun. 2017;8:1436.
Kasson P, DiMaio F, Yu X, Lucas-Staat S, Krupovic M, Schouten S, et al. Model for a novel membrane envelope in a filamentous hyperthermophilic virus. Elife. 2017;6:e26268.
DiMaio F, Yu X, Rensen E, Krupovic M, Prangishvili D, Egelman EH. Virology. A virus that infects a hyperthermophile encapsidates A-form DNA. Science. 2015;348:914–7.
Hong C, Pietila MK, Fu CJ, Schmid MF, Bamford DH, Chiu W. Lemon-shaped halo archaeal virus His1 with uniform tail but variable capsid structure. Proc Natl Acad Sci USA. 2015;112:2449–54.
Stedman KM, DeYoung M, Saha M, Sherman MB, Morais MC. Structural insights into the architecture of the hyperthermophilic Fusellovirus SSV1. Virology. 2015;474:105–9.
Krupovic M, Cvirkaite-Krupovic V, Iranzo J, Prangishvili D, Koonin EV. Viruses of archaea: Structural, functional, environmental and evolutionary genomics. Virus Res. 2018;244:181–93.
Iranzo J, Krupovic M, Koonin EV. The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. MBio. 2016;7:e00978–16.
Iranzo J, Koonin EV, Prangishvili D, Krupovic M. Bipartite network analysis of the Archaeal Virosphere: evolutionary connections between viruses and capsidless mobile elements. J Virol. 2016;90:11043–55.
Quemin ER, Chlanda P, Sachse M, Forterre P, Prangishvili D, Krupovic M. Eukaryotic-like virus budding in archaea. MBio. 2016;7:e01439–16.
Bize A, Karlsson EA, Ekefjard K, Quax TE, Pina M, Prevost MC, et al. A unique virus release mechanism in the Archaea. Proc Natl Acad Sci USA. 2009;106:11306–11.
Brumfield SK, Ortmann AC, Ruigrok V, Suci P, Douglas T, Young MJ. Particle assembly and ultrastructural features associated with replication of the lytic archaeal virus sulfolobus turreted icosahedral virus. J Virol. 2009;83:5964–70.
Quax TE, Krupovic M, Lucas S, Forterre P, Prangishvili D. The Sulfolobus rod-shaped virus 2 encodes a prominent structural component of the unique virion release system in Archaea. Virology. 2010;404:1–4.
Snyder JC, Brumfield SK, Peng N, She Q, Young MJ. Sulfolobus turreted icosahedral virus c92 protein responsible for the formation of pyramid-like cellular lysis structures. J Virol. 2011;85:6287–92.
Krupovic M, White MF, Forterre P, Prangishvili D. Postcards from the edge: structural genomics of archaeal viruses. Adv Virus Res. 2012;82:33–62.
Quemin ER, Pietila MK, Oksanen HM, Forterre P, Rijpstra WI, Schouten S, et al. Sulfolobus spindle-shaped virus 1 contains glycosylated capsid proteins, a cellular chromatin protein, and host-derived lipids. J Virol. 2015;89:11681–91.
Rensen EI, Mochizuki T, Quemin E, Schouten S, Krupovic M, Prangishvili D. A virus of hyperthermophilic archaea with a unique architecture among DNA viruses. Proc Natl Acad Sci USA. 2016;113:2478–83.
Wang F, Cvirkaite-Krupovic V, Kreutzberger MAB, Su Z, de Oliveira GAP, Osinski T, et al. An extensively glycosylated archaeal pilus survives extreme conditions. Nat Microbiol. 2019;4:1401–10.
Iranzo J, Lobkovsky AE, Wolf YI, Koonin EV. Evolutionary dynamics of the prokaryotic adaptive immunity system CRISPR-Cas in an explicit ecological context. J Bacteriol. 2013;195:3834–44.
Athukoralage JS, McMahon SA, Zhang C, Grüschow S, Graham S, Krupovic M, et al. A viral ring nuclease anti-CRISPR subverts type III CRISPR immunity. Nature. 2020;577:572–5.
Bhoobalan-Chitty Y, Johansen TB, Di Cianni N, Peng X. Inhibition of type III CRISPR-Cas immunity by an archaeal virus-encoded anti-CRISPR protein. Cell. 2019;179:448–58 e11.
He F, Bhoobalan-Chitty Y, Van LB, Kjeldsen AL, Dedola M, Makarova KS, et al. Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. Nat Microbiol. 2018;3:461–9.
Hurwitz S, Lowenstern JB. Dynamics of the Yellowstone hydrothermal system. Rev Geophys. 2014;52:375–411.
Munson-McGee JH, Peng S, Dewerff S, Stepanauskas R, Whitaker RJ, Weitz JS, et al. A virus or more in (nearly) every cell: ubiquitous networks of virus-host interactions in extreme environments. ISME J. 2018;12:1706–14.
Snyder JC, Bateson MM, Lavin M, Young MJ. Use of cellular CRISPR (clusters of regularly interspaced short palindromic repeats) spacer-based microarrays for detection of viruses in environmental samples. Appl Environ Microbiol. 2010;76:7251–8.
Bolduc B, Wirth JF, Mazurie A, Young MJ. Viral assemblage composition in Yellowstone acidic hot springs assessed by network analysis. ISME J. 2015;9:2162–77.
Piochi M, Kilburn CRJ, Di Vito MA, Mormone A, Tramelli A, Troise C, et al. The volcanic and geothermally active Campi Flegrei caldera: an integrated multidisciplinary image of its buried structure. Int Journ Earth Sci. 2014;103:401–21.
Piochi M, Mormone A, Balassone G, Strauss H, Troise C, De Natale G. Native sulfur, sulfates and sulfides from the active Campi Flegrei volcano (southern Italy): Genetic environments and degassing dynamics revealed by mineralogy and isotope geochemistry. J Volcano Geotherm Res. 2015;304:180–93.
Piochi M, Mormone A, Strauss H, Balassone G. The acid sulfate zone and the mineral alteration styles of the Roman Puteoli (Neapolitan area, Italy): clues on fluid fracturing progression at the Campi Flegrei volcano. Solid Earth. 2019;10:1809–31.
Menzel P, Gudbergsdottir SR, Rike AG, Lin L, Zhang Q, Contursi P, et al. Comparative metagenomics of eight geographically remote terrestrial hot springs. Micro Ecol. 2015;70:411–24.
Ciniglia C, Yoon HS, Pollio A, Pinto G, Bhattacharya D. Hidden biodiversity of the extremophilic Cyanidiales red algae. Mol Ecol. 2004;13:1827–38.
Gudbergsdóttir SR, Menzel P, Krogh A, Young M, Peng X. Novel viral genomes identified from six metagenomes reveal wide distribution of archaeal viruses and high viral diversity in terrestrial hot springs. Environ Microbiol. 2016;18:863–74.
Häring M, Peng X, Brugger K, Rachel R, Stetter KO, Garrett RA, et al. Morphology and genome organization of the virus PSV of the hyperthermophilic archaeal genera Pyrobaculum and Thermoproteus: a novel virus family, the Globuloviridae. Virology. 2004;323:233–42.
Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, et al. Screening for Sulfolobales, their Plasmids and their Viruses in Icelandic Solfataras. Syst Appl Microbiol. 1993;16:609–28.
Quemin ER, Lucas S, Daum B, Quax TE, Kuhlbrandt W, Forterre P, et al. First insights into the entry process of hyperthermophilic archaeal viruses. J Virol. 2013;87:13379–85.
Prangishvili D, Krupovic M, ICTV Report Consortium. ICTV virus taxonomy profile: Globuloviridae. J Gen Virol. 2018;99:1357–8.
Ahn DG, Kim SI, Rhee JK, Kim KP, Pan JG, Oh JW. TTSV1, a new virus-like particle isolated from the hyperthermophilic crenarchaeote Thermoproteus tenax. Virology. 2006;351:280–90.
Prangishvili D, Rensen E, Mochizuki T, Krupovic M, ICTV Report Consortium. ICTV Virus Taxonomy Profile: Tristromaviridae. J Gen Virol. 2019;100:135–6.
Neumann H, Schwass V, Eckerskorn C, Zillig W. Identification and characterization of the genes encoding three structural proteins of the Thermoproteus tenax virus TTV1. Mol Gen Genet. 1989;217:105–10.
Prangishvili D, Koonin EV, Krupovic M. Genomics and biology of Rudiviruses, a model for the study of virus-host interactions in Archaea. Biochem Soc Trans. 2013;41:443–50.
Bautista MA, Black JA, Youngblut ND, Whitaker RJ. Differentiation and Structure in Sulfolobus islandicus Rod-Shaped Virus Populations. Viruses. 2017;9:E120.
Meier-Kolthoff JP, Goker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33:3396–404.
Liu Y, Brandt D, Ishino S, Ishino Y, Koonin EV, Kalinowski J, et al. New archaeal viruses discovered by metagenomic analysis of viral communities in enrichment cultures. Environ Microbiol. 2019;21:2002–14.
Roux S, Krupovic M, Daly RA, Borges AL, Nayfach S, Schulz F, et al. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes. Nat Microbiol. 2019;4:1895–906.
Roux S, Adriaenssens EM, Dutilh BE, Koonin EV, Kropinski AM, Krupovic M, et al. Minimum Information about an Uncultivated Virus Genome (MIUViG). Nat Biotechnol. 2019;37:29–37.
Edwards RA, McNair K, Faust K, Raes J, Dutilh BE. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol Rev. 2016;40:258–72.
Pourcel C, Touchon M, Villeriot N, Vernadet JP, Couvin D, Toffano-Nioche C, et al. CRISPRCasdb a successor of CRISPRdb containing CRISPR arrays and cas genes from complete genome sequences, and tools to download and query lists of repeats and spacers. Nucleic Acids Res. 2020;48:D535–44.
Ågren J, Sundström A, Håfström T, Segerman B. Gegenees: fragmented alignment of multiple genomes for determining phylogenomic distances and genetic signatures unique for specified target groups. PLoS ONE. 2012;7:e39107.
Simmonds P, Adams MJ, Benko M, Breitbart M, Brister JR, Carstens EB, et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat Rev Microbiol. 2017;15:161–8.
Häring M, Rachel R, Peng X, Garrett RA, Prangishvili D. Viral diversity in hot springs of Pozzuoli, Italy, and characterization of a unique archaeal virus, Acidianus bottle-shaped virus, from a new family, the Ampullaviridae. J Virol. 2005;79:9904–11.
Vestergaard G, Haring M, Peng X, Rachel R, Garrett RA, Prangishvili D. A novel rudivirus, ARV1, of the hyperthermophilic archaeal genus Acidianus. Virology. 2005;336:83–92.
Prangishvili D, Vestergaard G, Haring M, Aramayo R, Basta T, Rachel R, et al. Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle. J Mol Biol. 2006;359:1203–16.
Vestergaard G, Aramayo R, Basta T, Haring M, Peng X, Brugger K, et al. Structure of the acidianus filamentous virus 3 and comparative genomics of related archaeal lipothrixviruses. J Virol. 2008;82:371–81.
Arnold HP, Ziese U, Zillig W. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology. 2000;272:409–16.
Mochizuki T, Sako Y, Prangishvili D. Provirus induction in hyperthermophilic archaea: characterization of Aeropyrum pernix spindle-shaped virus 1 and Aeropyrum pernix ovoid virus 1. J Bacteriol. 2011;193:5412–9.
Wiedenheft B, Stedman K, Roberto F, Willits D, Gleske AK, Zoeller L, et al. Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J Virol. 2004;78:1954–61.
Pauly MD, Bautista MA, Black JA, Whitaker RJ. Diversified local CRISPR-Cas immunity to viruses of Sulfolobus islandicus. Philos Trans R Soc Lond B Biol Sci. 2019;374:20180093.
Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol. 2009;11:457–66.
Stedman KM, Clore A, Combet-Blanc Y (2006). Biogeographical diversity of archaeal viruses. In: Logan NA, Lappin-Scott HM, Oyston PCF (eds). Prokaryotic Diversity: Mechanisms and Significance. Cambridge University Press: Cambridge. pp 131–43.
Snyder JC, Wiedenheft B, Lavin M, Roberto FF, Spuhler J, Ortmann AC, et al. Virus movement maintains local virus population diversity. Proc Natl Acad Sci USA. 2007;104:19102–7.
Medvedeva S, Liu Y, Koonin EV, Severinov K, Prangishvili D, Krupovic M. Virus-borne mini-CRISPR arrays are involved in interviral conflicts. Nat Commun. 2019;10:5204.
Atanasova NS, Roine E, Oren A, Bamford DH, Oksanen HM. Global network of specific virus-host interactions in hypersaline environments. Environ Microbiol. 2012;14:426–40.
Atanasova NS, Bamford DH, Oksanen HM. Virus-host interplay in high salt environments. Environ Microbiol Rep. 2016;8:431–44.
Laidler JR, Shugart JA, Cady SL, Bahjat KS, Stedman KM. Reversible inactivation and desiccation tolerance of silicified viruses. J Virol. 2013;87:13927–9.
De Sordi L, Khanna V, Debarbieux L. The gut microbiota facilitates drifts in the genetic diversity and infectivity of bacterial viruses. Cell Host Microbe. 2017;22:801–8, e3.
Paul BG, Bagby SC, Czornyj E, Arambula D, Handa S, Sczyrba A, et al. Targeted diversity generation by intraterrestrial archaea and archaeal viruses. Nat Commun. 2015;6:6585.
Klein R, Rossler N, Iro M, Scholz H, Witte A. Haloarchaeal myovirus phiCh1 harbours a phase variation system for the production of protein variants with distinct cell surface adhesion specificities. Mol Microbiol. 2012;83:137–50.
Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A, et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537:689–93.
Acknowledgements
This work was supported by l’Agence Nationale de la Recherche (France) project ENVIRA (to M.K.) and the European Union’s Horizon 2020 research and innovation program under grant agreement 685778, project VIRUS X (to D.P.). Y.L. is a recipient of the Pasteur-Roux-Cantarini Fellowship from Institut Pasteur. D.P.B. is part of the Pasteur—Paris University (PPU) International PhD Program, which has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 665807. We are also grateful to the Ultrastructural BioImaging (UTechS UBI) unit of Institut Pasteur for access to electron microscopes and Marc Monot from the Biomics Platform of Institut Pasteur for helpful discussions on genome assembly.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Baquero, D.P., Contursi, P., Piochi, M. et al. New virus isolates from Italian hydrothermal environments underscore the biogeographic pattern in archaeal virus communities. ISME J 14, 1821–1833 (2020). https://doi.org/10.1038/s41396-020-0653-z
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-020-0653-z
This article is cited by
-
Reply to: Natively expressed AcrIII-1 does not function as an anti-CRISPR protein
Nature (2025)
-
Evolutionary diversification of methanotrophic ANME-1 archaea and their expansive virome
Nature Microbiology (2023)
-
Archaeal extracellular vesicles are produced in an ESCRT-dependent manner and promote gene transfer and nutrient cycling in extreme environments
The ISME Journal (2021)
-
Lytic archaeal viruses infect abundant primary producers in Earth’s crust
Nature Communications (2021)