Abstract
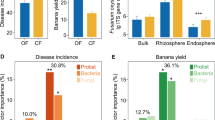
Soil-borne plant diseases are increasingly causing devastating losses in agricultural production. The development of a more refined model for disease prediction can aid in reducing crop losses through the use of preventative control measures or soil fallowing for a planting season. The emergence of high-throughput DNA sequencing technology has provided unprecedented insight into the microbial composition of diseased versus healthy soils. However, a single independent case study rarely yields a general conclusion predictive of the disease in a particular soil. Here, we attempt to account for the differences among various studies and plant varieties using a machine-learning approach based on 24 independent bacterial data sets comprising 758 samples and 22 independent fungal data sets comprising 279 samples of healthy or Fusarium wilt-diseased soils from eight different countries. We found that soil bacterial and fungal communities were both clearly separated between diseased and healthy soil samples that originated from six crops across nine countries or regions. Alpha diversity was consistently greater in the fungal community of healthy soils. While diseased soil microbiomes harbored higher abundances of Xanthomonadaceae, Bacillaceae, Gibberella, and Fusarium oxysporum, the healthy soil microbiome contained more Streptomyces Mirabilis, Bradyrhizobiaceae, Comamonadaceae, Mortierella, and nonpathogenic fungi of Fusarium. Furthermore, a random forest method identified 45 bacterial OTUs and 40 fungal OTUs that categorized the health status of the soil with an accuracy >80%. We conclude that these models can be applied to predict the potential for occurrence of F. oxysporum wilt by revealing key biological indicators and features common to the wilt-diseased soil microbiome.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Raw sequence data obtained in this study have been deposited in Genome Sequence Archive in the BIG Data Center, Chinese Academy of Sciences under accession codes CRA002340. All scripts for computational analysis and corresponding raw data are available at https://github.com/taowenmicro/Wen-etal-200214-paper-code.
References
Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48.
Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95 6578–83.
Gentile CL, Weir TL. The gut microbiota at the intersection of diet and human health. Science. 2018;362:776–80.
Penesyan A, Kjelleberg S, Egan S. Development of novel drugs from marine surface associated microorganisms. Mar drugs. 2010;8:438–59.
Chaparro JM, Sheflin AM, Manter DK, Vivanco JM. Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fertil Soils. 2012;48:489–99.
Mäder P, Fliessbach A, Dubois D, Gunst L, Fried P, Niggli U. Soil fertility and biodiversity in organic farming. Science. 2002;296:1694–7.
Classen AT, Sundqvist MK, Henning JA, Newman GS, Moore JA, Cregger MA, et al. Direct and indirect effects of climate change on soil microbial and soil microbial‐plant interactions: what lies ahead? Ecosphere. 2015;6:1–21.
de Vries FT, Griffiths RI, Bailey M, Craig H, Girlanda M, Gweon HS, et al. Soil bacterial networks are less stable under drought than fungal networks. Nat Commun. 2018;9:3033.
Van Der Heijden MG, Bardgett RD, Van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. 2008;11:296–310.
Hawkes CV, Wren IF, Herman DJ, Firestone MK. Plant invasion alters nitrogen cycling by modifying the soil nitrifying community. Ecol Lett. 2005;8:976–85.
Yuan J, Zhao J, Wen T, Zhao M, Li R, Goossens P, et al. Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome. 2018;6:156.
De Corato U, Patruno L, Avella N, Lacolla G, Cucci G. Composts from green sources show an increased suppressiveness to soilborne plant pathogenic fungi: Relationships between physicochemical properties, disease suppression, and the microbiome. Crop Prot. 2019;124:104870.
Finkel OM, Castrillo G, Paredes SH, González IS, Dangl JL. Understanding and exploiting plant beneficial microbes. Curr Opin Plant Biol. 2017;38:155–63.
Berendsen RL, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends plant Sci. 2012;17:478–86.
Ploetz RC. Fusarium wilt of banana. Phytopathology. 2015;105:1512–21.
Cha J-Y, Han S, Hong H-J, Cho H, Kim D, Kwon Y, et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 2016;10:119.
Gordon TR. Fusarium oxysporum and the Fusarium Wilt Syndrome. Annu Rev Phytopathol. 2017;55:23–39.
Laurence MH, Burgess LW, Summerell BA, Liew ECY. High levels of diversity in Fusarium oxysporum from non-cultivated ecosystems in Australia. Fungal Biol. 2012;116:289–97.
Nyvad B, Fejerskov O. An ultrastructural-study of bacterial invasion and tissue breakdown in human experimental root-surface caries. J Dent Res. 1990;69:1118–25.
Klein E, Ofek M, Katan J, Minz D, Gamliel A. Soil suppressiveness to Fusarium disease: shifts in root microbiome associated with reduction of pathogen root colonization. Phytopathology. 2012;103:23–33.
Mendes LW, Mendes R, Raaijmakers JM, Tsai SM. Breeding for soil-borne pathogen resistance impacts active rhizosphere microbiome of common bean. ISME J. 2018;12:3038–42.
Xiong W, Li R, Ren Y, Liu C, Zhao Q, Wu H, et al. Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biol Biochem. 2017;107:198–207.
Ye XF, Li ZK, Luo X, Wang WH, Li YK, Li R, et al. A predatory myxobacterium controls cucumber Fusarium wilt by regulating the soil microbial community. Microbiome. 2020;8:17.
Zhang S, Raza W, Yang X, Hu J, Huang Q, Xu Y, et al. Control of Fusarium wilt disease of cucumber plants with the application of a bioorganic fertilizer. Biol Fertil Soils. 2008;44:1073.
Fu L, Penton CR, Ruan Y, Shen Z, Xue C, Li R, et al. Inducing the rhizosphere microbiome by biofertilizer application to suppress banana Fusarium wilt disease. Soil Biol Biochem. 2017;104:39–48.
Shen Z, Ruan Y, Xue C, Zhong S, Li R, Shen Q, et al. Soils naturally suppressive to banana Fusarium wilt disease harbor unique bacterial communities. Plant. 2015;393:21–33.
De Corato U, Patruno L, Avella N, Salimbeni R, Lacolla G, Cucci G, et al. Soil management under tomato-wheat rotation increases the suppressive response against Fusarium wilt and tomato shoot growth by changing the microbial composition and chemical parameters. Appl Soil Ecol. 2020;154:103601.
Zhou D, Jing T, Chen Y, Wang F, Qi D, Feng R, et al. Deciphering microbial diversity associated with Fusarium wilt-diseased and disease-free banana rhizosphere soil. BMC Microbiol. 2019;19:161.
da C Jesus E, Marsh TL, Tiedje JM, de S Moreira FM. Changes in land use alter the structure of bacterial communities in Western Amazon soils. ISME J. 2009;3:1004–11.
Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–31.
Mercado-Blanco J, Abrantes I, Barra Caracciolo A, Bevivino A, Ciancio A, Grenni P, et al. Belowground microbiota and the health of tree crops. Front Microbiol. 2018;9:1006.
Ramirez KS, Knight CG, de Hollander M, Brearley FQ, Constantinides B, Cotton A, et al. Detecting macroecological patterns in bacterial communities across independent studies of global soils. Nat Microbiol. 2018;3:189–96.
Qiu M, Zhang R, Xue C, Zhang S, Li S, Zhang N, et al. Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol Fertil Soils. 2012;48:807–16.
Wang B, Li R, Ruan Y, Ou Y, Zhao Y, Shen Q. Pineapple–banana rotation reduced the amount of Fusarium oxysporum more than maize–banana rotation mainly through modulating fungal communities. Soil Biol Biochem. 2015b;86:77–86.
Alabouvette C. Fusarium wilt suppressive soils: an example of disease-suppressive soils. Australas Plant Pathol. 1999;28:57–64.
Hornby D. Suppressive soils. Australas Plant Pathol. 1983;21:65–85.
Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335.
Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584.
McDonald D, Clemente JC, Kuczynski J, Rideout JR, Stombaugh J, Wendel D, et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. GigaScience. 2012;1:7.
Liaw A, Wiener M. Classification and regression by randomForest. R N. 2002;2:18–22.
Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–97.
Wright RE. Logistic regression. In Grimm LG, & Yarnold PR, (Eds), Reading and understanding multivariate statistics (p. 217–244). Washington, DC: American Psychological Association; 1995.
Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc. 1996;58:267–88.
Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–1.
Statnikov A, Henaff M, Narendra V, Konganti K, Li Z, Yang L, et al. A comprehensive evaluation of multicategory classification methods for microbiomic data. Microbiome. 2013;1:11.
Zhang J, Liu Y-X, Zhang N, Hu B, Jin T, Xu H, et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat Biotechnol. 2019;37:676–84.
Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8:e1002687.
Csardi G, Nepusz T. The igraph software package for complex network research. Int J Complex Syst. 2006;1695:1–9.
Newman ME. The structure and function of complex networks. SIAM Rev. 2003;45:167–56.
Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. Msystems. 2016;1:e00009–15.
McKay G, Brown AE, Bjourson A, Mercer P. Molecular characterisation of Alternaria linicola and its detection in linseed. Eur J Plant Pathol. 1999;105:157–66.
Adams RI, Bateman AC, Bik HM, Meadow JF. Microbiota of the indoor environment: a meta-analysis. Microbiome. 2015;3:49.
Cornejo-Granados F, Gallardo-Becerra L, Leonardo-Reza M, Ochoa-Romo JP, Ochoa-Leyva A. A meta-analysis reveals the environmental and host factors shaping the structure and function of the shrimp microbiota. PeerJ. 2018;6:e5382.
Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, et al. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol. 2013;9:e1002863.
Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun. 2017;8:1784.
Rocca JD, Simonin M, Blaszczak JR, Ernakovich JG, Gibbons SM, Midani FS, et al. The Microbiome Stress Project: towards a global meta-analysis of environmental stressors and their effects on microbial communities. Front Microbiol. 2018;9:3272.
Gonzalez A, Navas-Molina JA, Kosciolek T, McDonald D, Vázquez-Baeza Y, Ackermann G, et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods. 2018;15:796–8.
Baxter NT, Ruffin MT, Rogers MA, Schloss PD. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016;8:37.
Belk A, Xu ZZ, Carter DO, Lynne A, Bucheli S, Knight R, et al. Microbiome data accurately predicts the postmortem interval using random forest regression models. Genes. 2018;9:104.
Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. 2017;551:585–9.
Manici L, Caputo F. Fungal community diversity and soil health in intensive potato cropping systems of the east Po valley, northern Italy. Ann Appl Biol. 2009;155:245–58.
Ploetz RC. Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp. cubense. Phytopathology. 2006;96:653–6.
Shen Z, Xue C, Taylor PWJ, Ou Y, Wang B, Zhao Y, et al. Soil pre-fumigation could effectively improve the disease suppressiveness of biofertilizer to banana Fusarium wilt disease by reshaping the soil microbiome. Biol Fertil Soils. 2018;54:793–806.
Wang B, Li R, Ruan Y, Ou Y, Zhao Y, Shen Q. Pineapple–banana rotation reduced the amount of Fusarium oxysporum more than maize–banana rotation mainly through modulating fungal communities. Soil Biol Biochem. 2015a;86:77–86.
Wu X, Guo S, Jousset A, Zhao Q, Shen Q. Bio-fertilizer application induces soil suppressiveness against Fusarium wilt disease by reshaping the soil microbiome. Soil Biol Biochem. 2017;114:238–47.
Forsyth LM, Smith LJ, Aitken EA. Identification and characterization of non-pathogenic Fusarium oxysporum capable of increasing and decreasing Fusarium wilt severity. Mycological Res. 2006;110:929–35.
Lemanceau P, Alabouvette C. Biological control of fusarium diseases by fluorescent Pseudomonas and non-pathogenic Fusarium. Crop Prot. 1991;10:279–86.
Liu L, Kong J, Cui H, Zhang J, Wang F, Cai Z, et al. Relationships of decomposability and C/N ratio in different types of organic matter with suppression of Fusarium oxysporum and microbial communities during reductive soil disinfestation. Biol Control. 2016;101:103–13.
Pieretti I, Royer M, Barbe V, Carrere S, Koebnik R, Cociancich S, et al. The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC genomics. 2009;10:616.
Li X, Zhang YN, Ding C, Jia Z, He Z, Zhang T, et al. Declined soil suppressiveness to Fusarium oxysporum by rhizosphere microflora of cotton in soil sickness. Biol Fertil soils. 2015;51:935–46.
Wu L, Yang B, Li M, Chen J, Xiao Z, Wu H, et al. Modification of rhizosphere bacterial community structure and functional potentials to control pseudostellaria heterophylla replant disease. Plant Dis. 2019;104:25–34.
Shang Q, Yang G, Wang Y, Wu X, Zhao X, Hao H, et al. Illumina-based analysis of the rhizosphere microbial communities associated with healthy and wilted Lanzhou lily (Lilium davidii var. unicolor) plants grown in the field. World J Microbiol Biotechnol. 2016;32:95.
Abbasi S, Safaie N, Sadeghi A, Shamsbakhsh M. Streptomyces strains induce resistance to Fusarium oxysporum f. sp. lycopersici race 3 in tomato through different molecular mechanisms. Front Microbiol. 2019;10:1505.
Liotti RG, da Silva Figueiredo MI, Soares MA. Streptomyces griseocarneus R132 controls phytopathogens and promotes growth of pepper (Capsicum annuum). Biol Control. 2019;138:104065.
Tahvonen R. Microbial control of plant diseases with Streptomyces spp. 1. Eppo Bull. 2010;18:55–9.
Yang Y, Zhang S-W, Li K-t. Antagonistic activity and mechanism of an isolated Streptomyces corchorusii stain AUH-1 against phytopathogenic fungi. World J Microbiol Biotechnol. 2019;35:145.
Cha J-Y, Han S, Hong H-J, Cho H, Kim D, Kwon Y, et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 2015;10:119–29.
Acknowledgements
We thank Yongxin Liu (institute of genetics and developmental biology, Chinese Academy of Sciences) for the comments on this manuscript, and the WeChat subscription ID “meta-Genome” and “Micro-Bioinformatics and microflora” for the analysis methods. This study was financially supported by Natural Science Foundation of Jiangsu Province (BK20170724), Natural Science Foundation of China (31902107), Special Fund for Agro-scientific Research in the Public Interest: integrated management technology of crop wilt disease (No. 201503110), the Innovative Research Team Development Plan of the Ministry of Education of China (Grant No. IRT_17R56), and the Fundamental Research Funds for the Central Universities (Grant No. KYT201802, KYXK2020010, and KJQN202017). JY was supported by National Postdoctoral Program for Innovative Talents (BX201600075).
Author information
Authors and Affiliations
Contributions
JY and TW conducted all experiments, conceived the study, and wrote the paper; HZ and MZ collected sequencing data; QS conceived the study, supervised the study, and wrote the paper; CRP and LST provided critical comments on the study, and helped write the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yuan, J., Wen, T., Zhang, H. et al. Predicting disease occurrence with high accuracy based on soil macroecological patterns of Fusarium wilt. ISME J 14, 2936–2950 (2020). https://doi.org/10.1038/s41396-020-0720-5
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-020-0720-5
This article is cited by
-
Differential adaptability of active ammonia-oxidizing bacteria and archaea to nitrogen amendment and fusarium in monocropped banana soils
BMC Microbiology (2025)
-
Combined effects of sound and temperature on the composition and function of bacterial and fungal communities in loess
BMC Microbiology (2025)
-
General variation in the Fusarium wilt rhizosphere microbiome
Nature Communications (2025)
-
Production Systems and Age Influence Fecal Mycobiota Diversity and Composition in Swine
Microbial Ecology (2025)
-
Compartment-specific core microbiomes and functions in rice
Plant and Soil (2025)