Abstract
Major depressive disorder (MDD) is characterized by persistent low mood and anhedonia. Mitochondrial dysfunction is linked to MDD, but the mechanisms are unclear. In this study, transcriptomic analysis of MDD patients’ peripheral blood found three key genes: TFAM, SURF1, and NDUFB9. Single-cell transcriptomic analysis of the prefrontal cortex (PFC) in MDD patients identified seven cell types. Analysis showed strong interactions between excitatory and inhibitory neurons in the PFC, with the three genes mainly in inhibitory neurons and NDUFB9 having the highest expression. We then established a chronic unpredictable mild stress (CUMS) mouse model. CUMS exposure induced depressive-like behaviors in mice, as evidenced by decreased sucrose preference, increased immobility time in the forced swim, and reduced activity and frequency of entries into the central area in the open field. Moreover, CUMS-exposed mice exhibited mitochondrial dysfunction in the prefrontal cortex (PFC). Notably, the expressions of TFAM, SURF1, and NDUFB9 were decreased in the PFC of CUMS mice, with the most significant decrease observed in NDUFB9. Subsequently, the overexpression of NDUFB9 in CUMS-treated mice significantly alleviated depressive-like behaviors, restored mitochondrial function and reduced the death of inhibitory neurons. It also enhanced mitophagy by PINK1/Parkin pathway. Inhibiting autophagy and mitophagy confirmed mitophagy’s pivotal role in NDUFB9-mediated restoration. Co-IP and protein half-life assays revealed that NDUFB9 stabilizes PINK1, thereby promoting mitophagy. In conclusion, our findings reveal a novel role of NDUFB9 on alleviating depression-like behavior by enhancing mitophagy, suggesting that targeting NDUFB9 could offer a promising therapeutic strategy for MDD.
Similar content being viewed by others
Introduction
MDD is a prevalent mental health condition marked by persistent low mood and anhedonia, as well as disrupted sleep, weight fluctuations, attention issues, and altered energy levels [1]. These symptoms are closely tied to impairments in brain areas crucial for mood regulation, including the hippocampus, prefrontal cortex, and striatum [2,3,4]. The diagnosis and treatment of depression largely rely on subjective symptom evaluations, as no reliable objective biomarkers are available for clinical use [5]. Moreover, existing antidepressant medications often have limited efficacy and numerous side effects, placing significant physical, psychological, and economic burdens on patients [6]. Although considerable research has been conducted on the biological underpinnings of depression, the exact pathophysiological processes remain poorly understood. Thus, we need to thoroughly investigate the pathogenesis of neuropsychiatric diseases, including depression, and clarify their neurorestorative mechanisms [7, 8].
Emerging research have indicated that the pathogenesis of depression is becoming more closely associated with mitophagy and the resulting mitochondrial dysfunction [9]. Mitochondria, as essential organelles in eukaryotic cells, are crucial for several key physiological functions, such as ATP synthesis, ROS production and clearance, and the complex regulation of apoptosis throughout the cell cycle [10]. Studies have shown that mitochondrial dysfunction is potentially exacerbating depressive symptoms by impairing neuronal energy metabolism, altering ROS levels, releasing inflammatory factors, and disrupting the synthesis and signaling of neurotransmitters [11]. Mitophagy plays a vital role in ensuring mitochondrial functionality by eliminating impaired mitochondria, thus stopping their buildup and ensuring mitochondrial quality control [12]. It has also been identified as a potential target of certain antidepressants. For instance, fluoxetine has been shown to enhance mitophagy, which helps eliminate damaged mitochondria and lower mtROS, in turn reducing cell death [13, 14]. The increasing evidence from these findings underscores the importance of mitophagy signaling pathways in MDD.
This study investigates the potential biomarkers and underlying mechanisms of depression by conducting bioinformatics analysis on key differentially expressed genes in the transcriptomic data of patients with MDD. Additionally, single-cell transcriptomics were employed to investigate key brain cell types involved in depression. Subsequently, molecular biology experiments were conducted to validate the analytical results and determine the roles and mechanisms of key genes in depression. These findings aim to provide new insights for the clinical diagnosis and treatment of depression.
Methods
Data collection
Transcriptomic Training Set: Data on gene expression and associated phenotypic information were obtained from the GSE52790 dataset available in the GEO database [15]. The platform used was GPL17976, with sequencing samples derived from peripheral blood. The dataset includes 22 samples in total, comprising 12 control samples (Control) and 10 major depressive disorder samples (Case).
Transcriptomic Validation Set: Gene expression profiles along with phenotypic data were retrieved from the GSE38206 dataset in the GEO database [16]. The platform used was GPL13607, with sequencing samples derived from peripheral blood mononuclear cells. The dataset includes 36 samples in total, comprising 18 control samples (Control) and 18 major depressive disorder samples (Case).
Single-cell RNA sequencing (scRNA-seq): Single-cell sequencing data from the GSE144136 dataset were downloaded from the GEO database [17]. The samples were derived from the prefrontal cortex, including 34 samples in total: 17 control samples (Control) and 17 major depressive disorder samples (Case).
Mitochondrial function-related genes (MF-RGs): A total of 1,136 MF-RGs were downloaded from the MitoCarta3.0 database.
Sleep disorder-related genes (SD-RGs): A total of 2,891 SD-RGs were retrieved from GeneCards by searching the keyword ‘sleep disorders’ and filtering with a Relevance Score ≥ 5.
This study analyzes publicly available datasets from the GEO database. All data have been fully de-identified and do not contain any personal identification information. Therefore, no additional ethical approval is required for this study.
Bioinformatics analysis
Differential gene expression analysis (DEGs)
DEGs between the control group (Control) and the disease group (Case) was performed using the “limma” package in R. Data normalization and batch effect correction were conducted to ensure the accuracy of the results. A linear model was applied to detect differentially expressed genes, with a threshold for the adj.p.value set at < 0.05.
Functional enrichment analysis
To assess the functions of differentially expressed genes (DEGs), we used two widely employed databases: GO and KEGG. Using the clusterProfiler package in R, we annotated the DEGs by mapping them to GO categories, enabling a systematic exploration of their roles both within and outside the cell. We also mapped the genes to the KEGG pathway database to identify potential signaling pathways and metabolic routes. To evaluate enrichment significance, we applied either the hypergeometric test or Fisher’s exact test, detecting over-representation of DEGs in specific categories and pathways. To control for false positives, we applied the Benjamini-Hochberg method for multiple test correction, filtering for significantly enriched terms with an FDR < 0.05.
PPI network
PPI networks were constructed using the STRING database to identify key proteins associated with differentially expressed genes and their predicted interactions. Cytoscape software was used to visualize the PPI network, and topological analysis identified central nodes and key pathways involved in disease progression.
Machine learning for gene selection
To optimize the candidate gene list and enhance research accuracy, we applied three machine learning techniques for feature selection: Least Absolute Shrinkage and Selection Operator (LASSO), Support Vector Machine (SVM), and the Boruta algorithm. (1) LASSO Regression: LASSO is a regularization technique that identifies genes strongly associated with disease by incorporating a penalty term to reduce redundant variables, thus improving model generalizability and avoiding overfitting. (2) SVM: SVM classifies genes critical to disease development based on gene expression levels, ensuring that the selected subset is highly predictive. (3) Boruta Algorithm: Based on Random Forest, Boruta eliminates irrelevant features and identifies important genes by evaluating their significance against randomly generated features, making it ideal for high-dimensional data. Please refer to the supplementary material for detailed algorithms and parameters.
ROC curve analysis
In this study, we used the “pROC” package in R to perform ROC curve analysis, evaluating the potential of selected genes as diagnostic and prognostic biomarkers. We assessed the diagnostic accuracy of candidate genes using AUC, which quantifies predictive accuracy. AUC values range from 0 to 1, with values closer to 1 indicating high sensitivity and specificity, signifying the gene’s ability to effectively distinguish between diseased and healthy individuals.
Single-cell RNA sequencing analysis
We used the Seurat package in R for processing and clustering single-cell RNA sequencing data. First, we standardized and denoised the data to remove technical noise while preserving biological signals. Next, we performed principal component analysis (PCA) to reduce data complexity and identify potential cell populations. Finally, we used known cell type-specific marker genes and cross-referenced with databases (CellMarker and PanglaoDB) to identify the major cell types expressing the target genes.
Animals and groups
The animals used in the experiments were male C57BL/6 mice. The mice were 8 to 10 weeks old, weighing between 25 and 30 grams. The mice were randomly assigned to four groups to ensure comparability of experimental data: 1) Control-Vector: Mice receiving only vehicle treatment with no stress exposure. 2) Control-NDUFB9-OE: Mice overexpressing the NDUFB9 gene. 3) CUMS-Vector: Mice exposed to CUMS and receiving vehicle treatment. 4) CUMS-NDUFB9-OE: Mice exposed to CUMS and overexpressing the NDUFB9 gene. To further clarify the specific role of mitophagy in CUMS, two targeted inhibitors were used: 3-MA (HY-19312, MCE, 10 mg/kg/day) via intraperitoneal injection [18], and Mdivi-1 (HY-15886, MCE, 3 mg/kg/day), also administered by intraperitoneal injection [19]. Both inhibitors were administered for two weeks, starting four weeks after the initiation of CUMS. Further subgroups within the CUMS-NDUFB9-OE group were established: 1) CUMS-NDUFB9-OE: Mice overexpressing NDUFB9 under CUMS without inhibitor treatment; 2) CUMS-NDUFB9-OE + 3-MA: Mice treated with 3-MA; and 3) CUMS-NDUFB9-OE + Mdivi-1: Mice treated with Mdivi-1. The experimental timeline and the brain regions used in the study are shown in Fig. 1A.
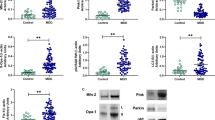
A, The experimental timeline and the brain regions used in the study; B–D, The mRNA level of TFAM, SURF1, and NDUFB9; E and F, The protein level of NDUFB9; G and H, Immunofluorescence results demonstrated the localization and expression of NDUFB9, scale bar = 100 μm; I, Sucrose preference percentage within 6 weeks; J, Immobility time in the FST; K, Total distance in the OFT; L, Number of crossings in the central area in the OFT; M and N, TUNEL assay results revealed inhibitory neuronal apoptosis in the PFC, scale bar = 100 μm. Data are presented as the mean ± SEM (n = 6 per group). *p < 0.05 and **p < 0.01 indicate significance compared to the control (Vector). #p < 0.05 and ##p < 0.01 indicate significance compared to the CUMS (Vector) group.
Stereotaxic delivery to the prefrontal cortex (PFC)
This study utilized adeno-associated virus (AAV) vectors provided by BrainVTA. During the injection procedure, the mice were first anesthetized. After anesthesia, the animals were placed in a stereotaxic apparatus for precise brain localization [20]. The injection coordinates were determined according to the standard brain atlas and were as follows: anterior-posterior (AP) = +2.3 mm, medial-lateral (ML) = ± 0.3 mm, and dorsal-ventral (DV) = − 2.0 mm [21]. A total of 500 nL of AAV vector was injected into each side of the cortical region. To ensure stable delivery of the vector, the injection speed was set to 40 nL/min. Three weeks post-injection, a CUMS model was established, and to further analyze the expression of NDUFB9, protein expression and mRNA levels of NDUFB9 in the PFC were quantified using Western blotting and PCR analysis (Supplementary Fig. 1A–C).
CUMS procedure
The CUMS was employed to induce depressive-like behaviors in mice, as described in previous studies (Supplementary Table 1) [22, 23]. To minimize external disturbances and ensure consistent environmental conditions, mice were individually housed in cages within a controlled room throughout the experiment. A series of stressors were randomly assigned to the animals, with one stressor being applied each day. To preserve the unpredictability of the stress exposure and prevent habituation, the specific stressor was varied randomly each week over a period of six consecutive weeks. The same stressor was not applied on consecutive days. The stressors included: restraint (1 h), swimming in water at either 4 °C or 45 °C (5 min), food or water deprivation (24 h), tail clamping (1 min), continuous nocturnal lighting (12 h), bedding dampened with 200 mL of water absorbed by 100 g of sawdust (24 h), and tilt of the cage at a 45° angle (24 h). The control group mice were handled identically to the CUMS group but were not exposed to any stressors.
Sucrose preference test (SPT)
The SPT is used to assess anhedonia in mice [24]. During the training phase, mice are given two bottles of 1% sucrose solution for 24 h to familiarize them with the substances. In the adaptation phase, one sucrose bottle is replaced with water. After 24 h of water deprivation to increase thirst, mice are given two bottles: one with sucrose solution and one with water. To avoid spatial bias, the bottles are switched every 12 h. Sucrose preference is calculated by dividing sucrose consumption by total fluid intake and multiplying by 100%. Lower preference scores indicate anhedonia, often linked to stress.
Forced swimming test (FST)
The FST was used to assess depressive-like behavior in mice. Each mouse was placed in a transparent cylinder filled with water (23–25 °C). The test began with an acclimation phase, followed by a 6-min swim session. After a 1-min adaptation period, the mouse’s immobility was measured for 5 min. Immobility was defined as minimal movement to keep the head above water or floating vertically. The test was recorded for later analysis, and fresh water was used between trials. Immobility time was analyzed to evaluate depressive-like behavior.
Open-field test (OFT)
The OFT was used to assess physical activity and anxiety-like behavior in mice. Each mouse was placed in the center of a square open field, and behavior was monitored using the Super Maze V2.0 video system. During a 5-min exploration period, the frequency of entries into the central area and the total walking distance were recorded. Fewer entries into the center indicate higher anxiety, while greater walking distance suggests lower anxiety and higher activity. All data were recorded for analysis to evaluate the effects of experimental conditions on mouse behavior.
qRT–PCR
Total RNA was extracted from mouse cortical tissue using TRIzol reagent and reverse transcribed into cDNA with the Top Green qPCR Kit. Quantitative PCR (qPCR) was performed using Gapdh mRNA as the internal reference gene for normalization. Relative expression levels of the target gene were calculated using the 2-ΔΔCq method. ΔCq values were determined by subtracting the Gapdh Cq from the target gene Cq, and ΔΔCq values were calculated by comparing experimental and control groups. The relative expression was then computed using the 2^(-ΔΔCq) formula. Primer sequences are listed in Supplementary Table 2.
TUNEL staining
Cell death was quantified using the TUNEL assay (Servicebio, China) according to the manufacturer’s instructions. Tissue samples were treated with proteinase K at 37 °C for 22 min and 0.1% Triton X-100 at room temperature for 20 min to increase permeability. The samples were then incubated with a 1:5 dilution of TUNEL Reagents 1 and 2 at 37 °C for 2 h to label fragmented DNA. DAPI staining was used for nuclear visualization. Samples were observed under a Leica fluorescence microscope, and images were analyzed with ImageJ software to quantify cell death.
Western blotting (WB) and Co-immunoprecipitation (Co-IP)
C57BL/6J mice were anesthetized with 2% isoflurane, and cortical tissue was collected using a brain mold to minimize damage. Protein was extracted from the cortex using a KeyGEN Biotech kit (China). Protein samples (30 μg each) were denatured, loaded onto an SDS-PAGE gel, separated, and transferred to a membrane. The membrane was incubated overnight at 4 °C with primary antibodies: anti-NDUFB9 (1:1000; Abcam, UK), anti-TFAM (1:1000; Abcam, UK), anti-NDUFS1 (1:1000; Proteintech, USA), anti-SDHB (1:1000; Proteintech, USA), anti-UQCRFS1 (1:1000; Proteintech, USA), anti-COX5B (1:1000; Proteintech, USA), anti-ATP5A (1:1000; Proteintech, USA), anti-PCG-1α (1:1000; Proteintech, USA), anti-NRF1 (1:1000; Proteintech, USA), anti-Beclin (1:1000; Proteintech, USA), anti-LC3 (1:1000; Proteintech, USA), anti-PINK1 (1:1000; Proteintech, USA), anti-Parkin (1:1000; Proteintech, USA), anti-P62 (1:1000; Wanleibio, China), and anti-β-actin (1:3000; Proteintech, USA). The membrane was then incubated with secondary antibodies (goat anti-rabbit or anti-mouse IgG from ZSJQ, China, 1:10,000) at room temperature for 1 h. Protein expression was normalized to β-actin levels. The protein bands were visualized using chemiluminescence and quantified with ImageJ software (NIH, USA).
For the Co-IP assay, cell proteins were extracted with RIPA buffer, and protein concentration was measured using the BCA assay. The lysates were incubated with specific antibodies (anti-NDUFB9, anti-PINK1, anti-IgG) and captured with Protein A/G agarose beads. After washing with PBS to remove non-specific binding, the precipitated proteins were analyzed by Western blot using the same primary antibodies as the initial analysis to confirm protein interactions.
Flow cytometric analysis
Mouse cortical cells were stained with JC-1 solution (C2006-1, Beyotime, China) and incubated at 37 °C for 30 min to 1 h. JC-1 aggregates in high membrane potential mitochondria, emitting red fluorescence, and remains in a monomeric form in low membrane potential mitochondria, emitting green fluorescence. After incubation, cells were washed with PBS and resuspended. Flow cytometry was used to excite JC-1 at 488 nm, with emissions detected at FL1 (525 ± 20 nm) for green fluorescence (low membrane potential) and FL2 (585 ± 20 nm) for red fluorescence (high membrane potential). Data were analyzed using FlowJo software to assess mitochondrial membrane potential.
Immunofluorescence staining
Brain tissue was fixed in 4% paraformaldehyde for 24 h and sectioned into 30 μm slices. After blocking with 10% goat serum for 1 h, the slices were incubated overnight at 4 °C with primary antibodies: anti-CAD65(1:200; Abcam, UK), anti-NDUFB9 (1:200; Abcam, UK), anti-LC3 (1:200; Proteintech, USA), and anti-TOMM20 (1:200; Proteintech, USA). Following three 5-min washes with PBS, the slices were incubated with secondary antibodies (Cy3-goat anti-rabbit IgG and FITC-goat anti-mouse IgG, 1:200; Proteintech, USA) for 2 h at room temperature. DAPI staining was used to visualize nuclei. Fluorescent images were captured with a Leica microscope and analyzed using ImageJ. Fluorescence images were converted to binary images, and thresholding was applied to identify positive staining regions. The region of interest (ROI) within the cortical area was defined based on anatomical markers, and quantitative analysis was performed by counting the number of double-labeled cells per square millimeter.
ATP assay
ATP levels were measured using the ATP detection kit (S0026, Beyotime) following the manufacturer’s instructions. Tissue samples were homogenized in lysis buffer, and protein concentration was determined by the BCA method. A 100 μl protein sample was mixed with 100 μl ATP detection buffer and immediately analyzed for luminescence using a luminometer. ATP concentration was quantified by comparing the luminescence signal to a standard curve and expressed as umol ATP/g protein.
DHE staining
This study uses the dihydroethidium (DHE) staining method to assess reactive oxygen species (ROS) levels [25]. Brain tissue samples were frozen and sectioned into 30 μm slices, then incubated with 5 μM DHE dye for 1 h at 37 °C in the dark. After three washes with PBS, the sections were mounted on slides. Fluorescent images were captured using a Leica fluorescence microscope, and ROS accumulation was quantified using ImageJ software.
Cell culture
SH-SY5Y cells (KeyGEN Biotech, China), derived from human neuroblastoma, were cultured in DMEM/F12 with 15% FBS for growth and differentiation. The cells were used between passages 3–20, and 1% penicillin-streptomycin to prevent contamination. The cells were incubated at 37 °C with 5% CO2 in a humidified incubator.
Molecular docking
First, the protein structures of NDUFB9 and PINK1 were obtained from the Uniprot database. Subsequently, the Hdock software was used to the interaction patterns between NDUFB9 and PINK1. Finally, the molecular docking results and their interaction patterns were analyzed using PyMOL 2.3.0 software (Schrödinger, USA).
Statistical analysis
The data were analyzed using GraphPad Prism version 9.0 (GraphPad Software, La Jolla, CA, USA). Results are presented as mean ± SEM from at least three independent experiments. For two-group comparisons, a Student’s t-test was used. For multiple-group comparisons, one-way ANOVA was performed, followed by Tukey’s post hoc test if a significant difference was found. A p-value of < 0.05 was considered significant. The statistical analysis in bioinformatics was referred to the supplementary material.
Results
Overexpression of NDUFB9 alleviates depression-like behaviors in CUMS mice
In this study, transcriptomic analysis of peripheral blood from MDD patients, along with data on genes related to sleep disorders (SD) and mitochondrial function, identified three key genes: TFAM (Transcription Factor A, Mitochondrial), SURF1 (SURF1 Cytochrome C Oxidase Assembly Factor), and NDUFB9 (NADH: Ubiquinone Oxidoreductase Subunit B9) (Supplementary Figure 2 and 3; Supplementary Table 3). Additionally, single-cell transcriptomic analysis of the PFC in MDD patients identified seven cell types. Analysis showed strong interactions between excitatory and inhibitory neurons in the PFC, with the three genes mainly in inhibitory neurons and NDUFB9 having the highest expression (Supplementary Figure 4). Subsequently, we assessed the relationship between CUMS-induced depressive-like behaviors and key genes using various experimental methods. The mRNA expression of TFAM, SURF1, and NDUFB9 is significantly decreased in the CUMS group (Fig. 1B–D, p < 0.01). Western Blot analysis further confirmed that the levels of NDUFB9 were significantly reduced in the CUMS group (Fig. 1E, F, p < 0.01), with a particularly notable decrease in inhibitory neurons (Fig. 1G, H, p < 0.01). The SPT showed that CUMS exposure significantly reduced sucrose preference (Fig. 1I, p < 0.001). However, overexpression of NDUFB9 reversed this effect (p < 0.01). The FST demonstrated a significant increase in the immobility time in the CUMS group (Fig. 1J, p < 0.01). NDUFB9 overexpression significantly reduced immobility time (p < 0.01). The OFT further supported these findings, with CUMS mice showing significantly reduced total walking distance (Fig. 1K, p < 0.01) and fewer entries into the center area (Fig. 1L, p < 0.01). NDUFB9 overexpression restored these behaviors to baseline levels (p < 0.01). Lastly, TUNEL staining demonstrated that NDUFB9 overexpression significantly reduced inhibitory neuronal apoptosis in the brains of CUMS mice, suggesting that NDUFB9 may protect neurons from stress-induced damage (Fig. 1M and N, p < 0.01).
Overexpression of NDUFB9 rescues mitochondrial dysfunction in the PFC of CUMS
We assessed the impact of NDUFB9 overexpression on mitochondrial function. In CUMS-exposed mice, the expression of mitochondrial electron transport chain complexes (I: NDUFS1, II: SDHB, III: UQCRFS1/RISP, IV: COX5B, V: ATP5A) was significantly reduced, indicating mitochondrial dysfunction. However, in mice overexpressing NDUFB9, the expression of these complexes was significantly restored (Fig. 2A–G, p < 0.01), suggesting that NDUFB9 overexpression can effectively reverse mitochondrial dysfunction. Further assessments of mitochondrial function also confirmed the protective effects of NDUFB9 overexpression. In CUMS-exposed mice, ATP levels in the prefrontal cortex (PFC) were markedly reduced. In contrast, ATP levels in NDUFB9-overexpressing mice were successfully restored (Fig. 2H, p < 0.01). Additionally, the measurement of mitochondrial membrane potential revealed a significant decrease in membrane potential in CUMS mice (Fig. 2I and J, p < 0.01). However, NDUFB9-overexpressing mice maintained a relatively high membrane potential, suggesting that NDUFB9 effectively preserves mitochondrial membrane integrity and function. ROS levels were significantly elevated in CUMS mice (Fig. 2K and L, p < 0.01). In contrast, ROS levels were significantly reduced in NDUFB9-overexpressing mice, indicating that NDUFB9 overexpression can alleviate oxidative stress and protect neuronal cells from damage.
A–G, The protein level of NDUFS1, SDHB, UQCRFS1/RISP, COX5B, and ATP5A in the PFC. H, Measurement of ATP production in the PFC; I and J, Flow cytometry was used to assess mitochondrial membrane potential; K and L, Measurement of ROS production in the PFC using the DHE assay, scale bar = 100 μm. Data are presented as the mean ± SEM (n = 6 per group). *p < 0.05 and **p < 0.01 indicate significance compared to the control (Vector). #p < 0.05 and ##p < 0.01 indicate significance compared to the CUMS (Vector) group.
Overexpression of NDUFB9 restores mitochondrial biogenesis and mitophagy in the PFC of CUMS
We investigated the protective mechanism of NDUFB9 in mitochondrial function under CUMS conditions in mice in this study. By evaluating mitochondrial dynamics proteins, we found no significant differences in the expression levels of mitochondrial fusion proteins (Mfn1, Mfn2, Opa1) and fission proteins (Drp1, Fis1) in the cortical tissue of CUMS-treated mice (Supplementary Fig. 5A–E, p > 0.05). Further analysis revealed that CUMS significantly reduced the expression of mitochondrial biogenesis related proteins PGC-1α, NRF1, and TFAM (Fig. 3A–D, p < 0.01). This suggests that stress may impair mitochondrial function by inhibiting mitochondrial biogenesis. However, overexpression of NDUFB9 effectively restored the expression of these proteins, indicating that NDUFB9 can protect mitochondrial function by promoting mitochondrial biogenesis.
A–D, The protein level of PCG-1α, NRF1, and TFAM in the PFC; E and F, Immunofluorescence results showed that LC3 and TOMM20 were co-located, scale bar = 100 μm; G-I, The protein level of PINK1, and Parkin in the PFC; J-M, The protein level of p62, Beclin, and LC3 in the PFC. Data are presented as the mean ± SEM (n = 6 per group). *p < 0.05 and **p < 0.01 indicate significance compared to the control (Vector). #p < 0.05 and ##p < 0.01 indicate significance compared to the CUMS (Vector) group.
The increase in mitochondrial biogenesis is consistent with the upregulation of mitochondrial gene expression and the increase in mitochondrial DNA (mtDNA) copy number. Therefore, we examined the levels of the outer mitochondrial membrane (OMM) protein TOMM20 and mtDNA copy number in the PFC. However, no significant differences were found among the four groups (Supplementary Figure 6A–C). The number of mitochondria within a cell, that is, mitochondrial content or mass, depends on the balance maintained between mitochondrial biogenesis and degradation (including processes such as mitophagy). The PINK1/Parkin-related pathway is the most traditional form of mitophagy. To further investigate mitochondrial clearance pathways, we first measured the levels of autophagy-related proteins P62, LC3, and Beclin. The results showed that CUMS treatment significantly increased the expression of P62, while the LC3II/LC3I ratio decreased, and Beclin expression was reduced, indicating that mitophagy was suppressed (Fig. 3J–M, p < 0.01). Notably, overexpression of NDUFB9 significantly restored the expression levels of these proteins, suggesting that NDUFB9 enhances mitophagy and promotes the clearance of damaged mitochondria, thereby improving mitochondrial quality control.
Immunofluorescence analysis showed that compared to the control group, the colocalization of LC3 (autophagy marker) and TOMM20 (mitochondrial marker) was significantly reduced in the cortical tissue of CUMS mice (Fig. 3E, F, p < 0.01). However, overexpression of NDUFB9 significantly increased the colocalization of these two markers (p < 0.01), further confirming that NDUFB9 promotes mitophagy.
Additionally, we assessed the PINK1/Parkin-mediated mitophagy pathway. We found that CUMS significantly decreased the protein levels of PINK1 and Parkin (Fig. 3G–I, p < 0.01). However, overexpression of NDUFB9 reversed this decline and restored the expression of PINK1 and Parkin, indicating that NDUFB9 regulates mitophagy through PINK1/Parkin pathway.
3-MA or Mdivi-1 inhibit the protective effects of NDUFB9 against CUMS-induced mitochondrial dysfunction
In this study, we used the autophagy inhibitor 3-MA and the mitophagy inhibitor Mdivi-1 to intervene in mice, immunofluorescence analysis revealed that they significantly reduced the colocalization of LC3 with TOMM20 (Fig. 4A, B, p < 0.01), indicating that the inhibitors effectively suppressed the mitophagy process. Further Western blot analysis showed that treatment with these inhibitors significantly decreased the protein expression levels of mitochondrial electron transport chain complexes I, II, III, IV, and ATP synthase in the PFC of the mice (Fig. 4C–I, p < 0.01).
A and B, Immunofluorescence results showed that LC3 and TOMM20 were co-located, scale bar = 100 μm; C-I, The protein level of NDUFS1, SDHB, UQCRFS1/RISP, COX5B, and ATP5A in the PFC; J, Measurement of ATP production in the PFC; K and L, Measurement of ROS production in the PFC using the DHE assay, scale bar = 100 μm; M and N, Flow cytometry was used to assess mitochondrial membrane potential. Data are presented as the mean ± SEM (n = 6 per group). *p < 0.05 and **p < 0.01 indicate significance compared to the NDUFB9-OE.
ATP measurements showed that treatment with 3-MA or Mdivi-1 effectively blocked the increase in ATP levels induced by NDUFB9 overexpression (Fig. 4J, p < 0.01). Likewise, mitochondrial membrane potential analysis revealed that the inhibitor group failed to restore the normal mitochondrial membrane potential induced by NDUFB9 overexpression (Fig. 4M, N, p < 0.01). Additionally, ROS measurements showed that 3-MA or Mdivi-1 suppressed the reduction in ROS levels induced by NDUFB9 overexpression (Fig. 4K, L, p < 0.01), which supports the involvement of mitophagy in NDUFB9’s antidepressant effects.
3-MA or Mdivi-1 inhibit the protective effects of NDUFB9 against CUMS-induced depression-like behavior
To determine the role of mitophagy in NDUFB9’s improvement of depressive-like behaviors, we first conducted behavioral tests. The results showed that after treatment with these two inhibitors, the protective effects of NDUFB9 overexpression on the SPT, FST, and OFT were abolished (Fig. 5A–D, p < 0.01). Further experiments also revealed that the protective effect of NDUFB9 on inhibitory neuronal apoptosis in the brain was lost (Fig. 5E and F, p < 0.01).
A, Sucrose preference percentage within 6 weeks; B, Immobility time in the FST; C, Total distance in the OFT; D, Number of crossings in the central area in the OFT; E and F, TUNEL assay results revealed inhibitory neuronal apoptosis in the PFC, scale bar = 100 μm; G, Protein interaction analysis using the String database; H, Molecular docking prediction, with PINK1 shown in red and NDUFB9 in blue; I, Co-IP analysis validating the interaction between PINK1 and NDUFB9 in neuronal cells; J, Westernblot analysis of the half-life of PINK1 protein in neuronal cells. Data are presented as the mean ± SEM (n = 6 per group). *p < 0.05 and **p < 0.01 indicate significance compared to the NDUFB9-OE.
NDUFB9 directly binds to PINK1 and stabilizes its expression in neuronal cells
To investigate the mechanism by which NDUFB9 regulates mitophagy, we first predicted the interaction proteins of NDUFB9 through PPI analysis, which identified PINK1 as one of the interacting proteins (Fig. 5G). Molecular docking prediction revealed that the binding affinity between NDUFB9 and PINK1 is -321.98 kcal/mol (Fig. 5H). Subsequently, we confirmed the direct interaction between NDUFB9 and PINK1 in neuronal cells through Co-IP experiments (Fig. 5I), indicating a potential impact on PINK1 stability. To further validate this, neuronal cells were treated with the protein synthesis inhibitor CHX. The results showed that NDUFB9 overexpression significantly prolonged the half-life of PINK1 protein (Fig. 5J, p < 0.01).
Discussion
Depression is a common mental disorder that has a significant impact on an individual’s physical and mental health. The pathological mechanisms of this complex disease remain unclear, and the clinical treatment outcomes are also inconsistent. This study investigates the potential role of NDUFB9 in MDD, with particular emphasis on its relationship to mitochondrial dysfunction. Our study shows that NDUFB9 plays a key role in mitochondrial function and improving depressive- like behaviors. It can stabilize PINK1, which activates the PINK1/Parkin pathway. This enhances mitophagy in neuronal cells. The increase in mitophagy improves the mitochondrial electron transport chain function. It also stabilizes the mitochondrial membrane potential, reduces ROS, and increases ATP levels. These mitochondrial improvements enhance neuronal function and significantly reduce depressive-like behaviors in CUMS mice (Fig. 6). Our findings offer valuable insights into the pathophysiological mechanisms underlying MDD.
NDUFB9 binds to PINK1 and inhibits its degradation, thereby activating the PINK1/Parkin pathway. This activation enhances neuronal mitophagy and improves mitochondrial function, including electron transport chain activity, membrane potential stabilization, reduced ROS levels, and increased ATP production. Consequently, these mitochondrial improvements enhance neuronal function and significantly mitigate depressive-like behaviors in CUMS (chronic unpredictable mild stress) mice.
Biomarkers in peripheral blood are closely associated with brain function. Research suggests that peripheral blood markers can reliably reflect brain health, playing a crucial role in neuroinjury, major depression, and neurodegenerative diseases [26,27,28]. Through bioinformatics analysis of peripheral blood RNA sequencing data from patients with depression, our study identified key genes (TFAM, SURF1, and NDUFB9) that are potentially involved in MDD, with a particular focus on mitochondrial function and sleep disorders. Research has shown that individuals with chronic sleep disorders (SD) are more likely to develop mental health conditions, including bipolar disorder, suicidal thoughts, and particularly MDD [29]. Furthermore, a bidirectional relationship may exist between MDD and SD: patients with depression are more prone to SD due to neurotransmitter abnormalities, while chronic sleep problems further exacerbate depressive symptoms [30, 31]. Meanwhile, mitochondrial function is also an important factor affecting the onset and progression of MDD. TFAM is a mitochondrial transcription factor that maintains the stability and replication of mitochondrial DNA (mtDNA). In the LPS-induced mouse model of depression-like behavior, the expression of TFAM is significantly reduced [19]. SURF1 encodes a protein essential for the assembly and proper functioning of cytochrome c oxidase (COX), a key enzyme in the mitochondrial electron transport chain. Mutations in SURF1 can lead to mitochondrial disorders, such as Leigh syndrome, by impairing COX assembly and activity [32]. NDUFB9 encodes a subunit of mitochondrial Complex I, a critical component of the electron transport chain involved in generating cellular energy through oxidative phosphorylation [33]. Research has found that NDUFB9 was reduced in the model of maternal deprivation combined with social isolation [34]. To gain a deeper understanding of how these genes impact brain function, we analyzed scRNA-seq from the brains of depression patients. Single-cell RNA sequencing is a powerful tool in brain research [35]. Compared to traditional bulk tissue analysis, single-cell RNA sequencing captures variability across different brain regions and cell types, yielding more precise data for understanding the molecular mechanisms underlying depression. Based on the single-cell RNA sequencing, seven cell types were annotated, including Astrocytes Endothelial cells, Excitatory neurons, Inhibitory neurons, Microglia/Macrophages, Oligodendrocytes, and Oligodendrocyte precursor cells. Our analysis shows that excitatory and inhibitory neurons coexist in the prefrontal cortex of patients with MDD, and they exhibit strong interactions, which aligns with the brain characteristics observed in depression [36, 37]. In particular, inhibitory neurons play a crucial role in the onset and progression of MDD [38]. The expression levels of the three key genes were higher in the control group compared to the MDD group, consistent with the results from the transcriptomic data, and all three genes are predominantly expressed in inhibitory neurons. Specifically, NDUFB9 shows the highest expression in inhibitory neurons. Next, we established a mouse model of depression using CUMS to validate the expression of these three genes. We found that the expression levels of the three genes were consistent with our analysis results. Moreover, NDUFB9 was the most differentially expressed gene in both the peripheral blood and the prefrontal cortex of patients with MDD, as well as in the prefrontal cortex of CUMS mice. Although no direct link between NDUFB9 and MDD has been reported, our analysis suggest that it may play a novel role in depression-like behaviors.
To investigate the role of NDUFB9 in MDD, we utilized a CUMS-induced mouse model. CUMS is the most commonly used, reliable, and effective rodent depression model, overcoming habituation and sex specificity [39]. The findings indicated that CUMS exposure led to depressive-like behaviors, as evidenced by a decrease in sucrose preference, increased immobility in the forced swim test, and reduced activity in the open field test. Furthermore, mice subjected to CUMS exhibited increased neuronal apoptosis in the brain. These findings are consistent with our earlier study [22]. Recently, mitochondrial dysfunction has attracted increasing attention in depression research. Several clinical and preclinical studies suggest that mitochondrial dysfunction, as a key regulator of cellular energy metabolism, plays a crucial role in the pathogenesis of depression [40]. Depressed individuals often exhibit significant mitochondrial abnormalities, particularly in brain regions essential for emotional regulation, such as the PFC [41]. The PFC has emerged as one of the regions most consistently impaired in MDD [42]. The neuron structure of the PFC region in patients with MDD has changed significantly, such as synapse reduction and dendrite complexity reduction [43]. Our findings indicate that CUMS-exposed mice exhibit mitochondrial dysfunction in the PFC, particularly in the mitochondrial electron transport chain, leading to a decrease in mitochondrial membrane potential, elevated ROS production, and impaired ATP synthesis. Moreover, overexpression of NDUFB9 in CUMS-treated mice significantly alleviated depressive-like behaviors and restored mitochondrial function. These results suggest that NDUFB9 may ameliorate CUMS-induced depressive-like behaviors by enhancing mitochondrial function.
To investigate the protective mechanism of NDUFB9 on mitochondria, we examined mitochondrial dynamics (including fission and fusion), mitochondrial biogenesis, and mitophagy. Our results indicated that NDUFB9 had no effect on mitochondrial dynamics-related proteins. The PGC-1α/NRF1/TFAM pathway plays a crucial role in regulating mitochondrial biogenesis [44]. Consistent with previous research reporting a reduction in the expression of PGC-1α, NRF1, and TFAM in the chronic mild stress model [45], our results demonstrated that NDUFB9 overexpression led to elevated levels of PGC-1α, NRF1, and TFAM, suggesting its involvement in enhancing mitochondrial biogenesis. Mitochondrial content or quality, which reflects the number of mitochondria within cells, is influenced by the balance between mitochondrial biogenesis and degradation, which includes processes like mitophagy [46]. In our study, CUMS exposure altered the expression levels of autophagy-related proteins, an effect that was reversed by the overexpression of NDUFB9. Additionally, NDUFB9 overexpression restored the expression of PINK1 and Parkin, two key proteins involved in the mitophagy pathway. These results suggest that NDUFB9 protects mitochondrial function primarily by promoting mitochondrial biogenesis and mitophagy.
To further explore the role of mitophagy in the antidepressant effects of NDUFB9, we utilized autophagy (3-MA) and mitophagy (Mdivi-1) inhibitors. 3-MA inhibits the formation of LC3-II and promotes p62 accumulation, thereby effectively blocking autophagosome formation [47]. Meanwhile, treatment with Mdivi-1 prevents the recruitment of PINK1 and Parkin to the mitochondria, thereby inhibiting the initiation of mitophagy [48]. Our findings demonstrated that inhibiting autophagy or mitophagy negated the effects of NDUFB9 in restoring mitochondrial function and alleviating depressive-like behavior. These results strongly support the involvement of mitophagy in the antidepressant mechanism of NDUFB9. Furthermore, our study revealed that NDUFB9 directly interacts with PINK1 and stabilizes its expression in neuronal cells. The primary mechanism of PINK1-mediated mitophagy involves the accumulation of full-length PINK1 on the outer mitochondrial membrane, where it recruits PRKN (Parkin RBR E3 ubiquitin protein ligase) and initiates the phosphorylation of ubiquitin chains [49]. On the inner mitochondrial membrane, PARL (presenilin-associated rhomboid-like protein) cleaves mature PINK1, leading to its destabilization. The cleaved PINK1 is subsequently degraded by the proteasome, resulting in the loss of its ability to trigger mitophagy [50]. Therefore, the interaction between NDUFB9 and PINK1 is likely crucial for sustaining the PINK1/Parkin-mediated mitophagy pathway and preventing mitochondrial dysfunction. These findings provide valuable insights into the molecular mechanisms underlying the antidepressant effects of NDUFB9.
This study has certain limitations. Firstly, the study used clinical samples for data analysis but only validated the results with the CUMS animal model. Although the CUMS model is the most commonly used in depression research, it still differs from clinical depression. In terms of symptom presentation, animals in the CUMS model mainly show behavioral changes, such as anhedonia in the sucrose preference test and increased immobility time in the forced swimming test. In contrast, human depression symptoms are more diverse, involving emotional, cognitive, and physiological aspects. Regarding the pathogenesis, the CUMS model is mainly related to stress - induced changes in the neuroendocrine and neurochemical systems, such as increased plasma corticosterone and decreased 5- HT levels. However, the pathogenesis of human depression is more complex, involving genetic, environmental, and psychological factors. Secondly, in the study of mitophagy, we only observed the most widely - studied PINK1/Parkin pathway and did not examine the BNIP3/NIX pathway. In future research, we will investigate the impact of NDUFB9 on the BNIP3/NIX pathway. Thirdly, the use of only male animals, while reducing hormonal variability, may restrict the translational relevance of our findings, as sex-specific mechanisms in depression remain to be explored.
In conclusion, our study provides evidence for a novel role of NDUFB9 in the pathophysiology of MDD. Our findings suggest that NDUFB9 may improve depressive-like behavior by promoting mitochondrial biogenesis and mitophagy, thereby restoring mitochondrial function. The interaction between NDUFB9 and PINK1 further supports the involvement of mitophagy in the antidepressant mechanism of NDUFB9. These findings contribute to a deeper understanding of the pathophysiological basis of MDD and provide potential therapeutic targets for the treatment of MDD.
Data availability
All the data in this research can be obtained from the corresponding author upon a justified request.
References
Jin Y, Cui R, Zhao L, Fan J, Li B. Mechanisms of Panax ginseng action as an antidepressant. Cell Prolif. 2019;52:e12696.
Tunc-Ozcan E, Peng CY, Zhu Y, Dunlop SR, Contractor A, Kessler JA. Activating newborn neurons suppresses depression and anxiety-like behaviors. Nat Commun. 2019;10:3768.
Belleau EL, Treadway MT, Pizzagalli DA. The impact of stress and major depressive disorder on hippocampal and medial prefrontal cortex morphology. Biol Psychiatry. 2019;85:443–53.
Bekhbat M, Li Z, Mehta ND, Treadway MT, Lucido MJ, Woolwine BJ, et al. Functional connectivity in reward circuitry and symptoms of anhedonia as therapeutic targets in depression with high inflammation: evidence from a dopamine challenge study. Mol Psychiatry. 2022;27:4113–21.
Carboni L, McCarthy DJ, Delafont B, Filosi M, Ivanchenko E, Ratti E, et al. Biomarkers for response in major depression: comparing paroxetine and venlafaxine from two randomised placebo-controlled clinical studies. Transl Psychiatry. 2019;9:182.
Wang Z, Meng Z, Chen C. Screening of potential biomarkers in peripheral blood of patients with depression based on weighted gene co-expression network analysis and machine learning algorithms. Front Psychiatry. 2022;13:1009911.
Huang H, Bach JR, Sharma HS, Chen L, Wu P, Sarnowska A, et al. The 2023 yearbook of Neurorestoratology. J Neurorestoratology. 2024;12:100136.
Huang H, Bach JR, Sharma HS, Saberi H, Jeon SR, Guo X, et al. The 2022 yearbook of Neurorestoratology. J Neurorestoratology. 2023;11:100054.
Tripathi A, Scaini G, Barichello T, Quevedo J, Pillai A. Mitophagy in depression: Pathophysiology and treatment targets. Mitochondrion. 2021;61:1–10.
Lee A, Hirabayashi Y, Kwon SK, Lewis TL Jr., Polleux F. Emerging roles of mitochondria in synaptic transmission and neurodegeneration. Curr Opin Physiol. 2018;3:82–93.
Zuo C, Cao H, Song Y, Gu Z, Huang Y, Yang Y, et al. Nrf2: An all-rounder in depression. Redox Biol. 2022;58:102522.
Kerr JS, Adriaanse BA, Greig NH, Mattson MP, Cader MZ, Bohr VA, et al. Mitophagy and Alzheimer’s disease: cellular and molecular mechanisms. Trends Neurosci. 2017;40:151–66.
Shu X, Sun Y, Sun X, Zhou Y, Bian Y, Shu Z, et al. The effect of fluoxetine on astrocyte autophagy flux and injured mitochondria clearance in a mouse model of depression. Cell Death Dis. 2019;10:577.
Wei Q, Zhou W, Zheng J, Li D, Wang M, Feng L, et al. Antidepressant effects of 3-(3,4-methylenedioxy-5-trifluoromethyl phenyl)-2E-propenoic acid isobutyl amide involve TSPO-mediated mitophagy signalling pathway. Basic Clin Pharmacol Toxicol. 2020;127:380–8.
Liu Z, Li X, Sun N, Xu Y, Meng Y, Yang C, et al. Microarray profiling and co-expression network analysis of circulating lncRNAs and mRNAs associated with major depressive disorder. PLoS One. 2014;9:e93388.
Belzeaux R, Bergon A, Jeanjean V, Loriod B, Formisano-Tréziny C, Verrier L, et al. Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl Psychiatry. 2012;2:e185.
Nagy C, Maitra M, Tanti A, Suderman M, Théroux JF, Davoli MA, et al. Single-nucleus transcriptomics of the prefrontal cortex in major depressive disorder implicates oligodendrocyte precursor cells and excitatory neurons. Nat Neurosci. 2020;23:771–81.
Su L, Lu H, Zhang D, Zhu X, Li J, Zong Y, et al. Total paeony glycoside relieves neuroinflammation to exert antidepressant effect via the interplay between NLRP3 inflammasome, pyroptosis and autophagy. Phytomedicine. 2024;128:155519.
Liu Q, Zhao JN, Fang ZT, Wang X, Zhang BG, He Y, et al. BGP-15 alleviates LPS-induced depression-like behavior by promoting mitophagy. Brain Behav Immun. 2024;119:648–64.
Sun Y, Cong T, Li L, Li J, Wu H, Chen X, et al. Baclofen attenuates cognitive deficits in post-cardiac arrest brain injury. Biochem Biophys Res Commun. 2022;602:135–41.
Zheng XX, Zhang CF, Li LQ, Ye JR, Ren SY, Zhang Z, et al. Improvement of astrocytic gap junction involves the anti-depressive effect of celecoxib through inhibition of NF-κB. Brain Res Bull. 2024;207:110871.
Sun Y, Zhao D, Song Q, Cong T, Li L, Wu H, et al. NMT2 alleviates depression-like behavior in a rat model of chronic unpredictable stress: An integrated proteomic and phosphoproteomic analysis. J Psychiatr Res. 2024;176:119–28.
Li H, Xiang Y, Zhu Z, Wang W, Jiang Z, Zhao M, et al. Rifaximin-mediated gut microbiota regulation modulates the function of microglia and protects against CUMS-induced depression-like behaviors in adolescent rat. J Neuroinflammation. 2021;18:254.
Yao D, Li R, Hao J, Huang H, Wang X, Ran L, et al. Melatonin alleviates depression-like behaviors and cognitive dysfunction in mice by regulating the circadian rhythm of AQP4 polarization. Transl Psychiatry. 2023;13:310.
Liu H, Li Y, Sun S, Xin Q, Liu S, Mu X, et al. Catalytically potent and selective clusterzymes for modulation of neuroinflammation through single-atom substitutions. Nat Commun. 2021;12:114.
Gonzalez-Ortiz F, Turton M, Kac PR, Smirnov D, Premi E, Ghidoni R, et al. Brain-derived tau: a novel blood-based biomarker for Alzheimer’s disease-type neurodegeneration. Brain. 2023;146:1152–65.
Ghaith HS, Nawar AA, Gabra MD, Abdelrahman ME, Nafady MH, Bahbah EI, et al. A literature review of traumatic brain injury biomarkers. Mol Neurobiol. 2022;59:4141–58.
Gadad BS, Jha MK, Czysz A, Furman JL, Mayes TL, Emslie MP, et al. Peripheral biomarkers of major depression and antidepressant treatment response: Current knowledge and future outlooks. J Affect Disord. 2018;233:3–14.
Morin CM, LeBlanc M, Daley M, Gregoire JP, Mérette C. Epidemiology of insomnia: prevalence, self-help treatments, consultations, and determinants of help-seeking behaviors. Sleep Med. 2006;7:123–30.
Fang H, Tu S, Sheng J, Shao A. Depression in sleep disturbance: a review on a bidirectional relationship, mechanisms and treatment. J Cell Mol Med. 2019;23:2324–32.
Yasugaki S, Okamura H, Kaneko A, Hayashi Y. Bidirectional relationship between sleep and depression. Neurosci Res. 2025;211:57–64.
Lee IC, Chiang KL. Clinical diagnosis and treatment of leigh syndrome based on SURF1: genotype and phenotype. Antioxidants (Basel). 2021;10:1950.
Li LD, Sun HF, Liu XX, Gao SP, Jiang HL, Hu X, et al. Down-regulation of NDUFB9 promotes breast cancer cell proliferation, metastasis by mediating mitochondrial metabolism. PLoS One. 2015;10:e0144441.
Vera-Montecinos A, Rodríguez-Mias R, MacDowell KS, García-Bueno B, Bris ÁG, Caso JR, et al. Analysis of molecular networks in the cerebellum in chronic schizophrenia: modulation by early postnatal life stressors in murine models. Int J Mol Sci. 2021;22:10076.
Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–42.
Corey F, Mounira B, Etienne SJBP. Somatostatin-positive gamma-aminobutyric acid interneuron deficits in depression: cortical microcircuit and therapeutic perspectives. Biol Psychiatry. 2017;82:549–59.
Chatzinakos C, Pernia CD, Morrison FG, Iatrou A, McCullough KM, Schuler H, et al. Single-nucleus transcriptome profiling of dorsolateral prefrontal cortex: mechanistic roles for neuronal gene expression, including the 17q21.31 Locus, in PTSD stress response. Am J Psychiatry. 2023;180:739–54.
Luscher B, Shen Q, Sahir N. The GABAergic deficit hypothesis of major depressive disorder. Mol Psychiatry. 2011;16:383–406.
Antoniuk S, Bijata M, Ponimaskin E, Wlodarczyk J. Chronic unpredictable mild stress for modeling depression in rodents: meta-analysis of model reliability. Neurosci Biobehav Rev. 2019;99:101–16.
Song Y, Cao H, Zuo C, Gu Z, Huang Y, Miao J, et al. Mitochondrial dysfunction: a fatal blow in depression. Biomed Pharmacother. 2023;167:115652.
Bansal Y, Kuhad A. Mitochondrial dysfunction in depression. Curr Neuropharmacol. 2016;14:610–8.
Pizzagalli DA, Roberts AC. Prefrontal cortex and depression. Neuropsychopharmacology. 2022;47:225–46.
Lin S, Huang L, Luo ZC, Li X, Jin SY, Du ZJ, et al. The ATP level in the medial prefrontal cortex regulates depressive-like behavior via the medial prefrontal cortex-lateral habenula pathway. Biol Psychiatry. 2022;92:179–92.
Shi M, Dong Z, Zhao K, He X, Sun Y, Ren J, et al. Novel insights into exhaustive exercise-induced myocardial injury: Focusing on mitochondrial quality control. Front Cardiovasc Med. 2022;9:1015639.
Khedr LH, Nassar NN, Rashed L, El-Denshary ED, Abdel-Tawab AM. TLR4 signaling modulation of PGC1-α mediated mitochondrial biogenesis in the LPS-Chronic mild stress model: Effect of fluoxetine and pentoxiyfylline. Life Sci. 2019;239:116869.
Tronstad KJ, Nooteboom M, Nilsson LI, Nikolaisen J, Sokolewicz M, Grefte S, et al. Regulation and quantification of cellular mitochondrial morphology and content. Curr Pharm Des. 2014;20:5634–52.
Shi Y, Tao M, Ma X, Hu Y, Huang G, Qiu A, et al. Delayed treatment with an autophagy inhibitor 3-MA alleviates the progression of hyperuricemic nephropathy. Cell Death Dis. 2020;11:467.
Wang H, Ye J, Peng Y, Ma W, Chen H, Sun H, et al. CKLF induces microglial activation via triggering defective mitophagy and mitochondrial dysfunction. Autophagy. 2024;20:590–613.
Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31.
Meissner C, Lorenz H, Hehn B, Lemberg MK. Intramembrane protease PARL defines a negative regulator of PINK1- and PARK2/Parkin-dependent mitophagy. Autophagy. 2015;11:1484–98.
Acknowledgements
We extend our gratitude to everyone who contributed to or supported this research endeavor.
Funding
This research received funding from the Liaoning Province Education Administration Foundation (The People’s Government of Liaoning Province) under Grant No: LJKZ0865. Additional support was provided through the “1 + X” Research Project conducted at the Second Hospital of Dalian Medical University.
Author information
Authors and Affiliations
Contributions
The literature search was conducted by Ye Sun, who also formulated the hypothesis, carried out the experiments, analyzed the results, and prepared the manuscript draft. Data analysis was supported by Liya Li, while Xianglong Yang was responsible for developing the animal model. Zhaoyang Xiao and Shengming Yin provided supervision and made significant revisions to the manuscript. All authors reviewed and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All experimental protocols were performed in strict compliance with the National Institutes of Health (NIH) Guidelines for the Care and Use of Laboratory Animals and were approved by the Animal Experiment Ethics Committee of Dalian Medical University (No: AEE19087).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, Y., Li, L., Yang, X. et al. NDUFB9 ameliorates CUMS-induced depression-like behavior by promoting mitophagy. Transl Psychiatry 15, 292 (2025). https://doi.org/10.1038/s41398-025-03502-4
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41398-025-03502-4