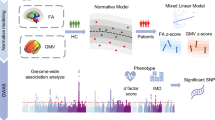
Abstract
Not everyone is equally likely to experience mental illness. What is the contribution of an individual’s genetic background and experiences of childhood adversity to that likelihood? And how do these risk factors interact at the level of the brain? This study explores these questions by investigating the relationship between genetic liability for mental illness, childhood adversity, and cortico-limbic connectivity in a large developmental sample drawn from the ABCD cohort (N = 6535). Canonical Correlation Analysis – a multivariate data-reduction technique – revealed two genetic dimensions of mental illness from the polygenic risk scores for ADHD, Anxiety, Depression, and Psychosis. The first dimension represented liability for broad psychopathology which was positively correlated with adversity. The second dimension represented neurodevelopmental-specific risk which negatively interacted with adversity, suggesting that neurodevelopmental symptoms may arise from unique combinations of genetic and environmental factors that differ from other symptom domains. Next, we investigated the cortico-limbic signature of adversity and genetic liability using Partial Least Squares. We found that the neural correlates of adversity broadly mirrored those of genetic liability, with adversity capturing most of the shared variance. These novel findings suggest that genetic and environmental risk overlap in the neural connections that underlie mental health symptomatology.
Similar content being viewed by others
Introduction
The prevalence of mental illness represents a significant global health challenge, with a myriad of factors contributing to its onset and progression [1]. Different combinations of interacting genetic, biological, and environmental constraints shape the neural substrates of mental health [2]. To better understand the etiology of common mental health conditions it is necessary to consider both genetic and environmental influences in combination and discern how they converge to produce risk-relevant neural and behavioural phenotypes [3,4,5].
The human genome interacts with the environmental factors to shape both physical and behavioural traits [6,7,8,9]. Genetic liability for mental illness is believed to manifest by altering the development of neural systems that play a role in symptom presentation, such as those involved in affective processing, fear learning, and social cognition [10,11,12,13]. The molecular embedding of adversity in stress-regulatory systems indicates that genetic and environmental risk may converge at the level of neurobiology [11, 13, 14] by altering stress-susceptible neural systems in similar ways [15,16,17]. Indeed, adversity exposed individuals often exhibit similar changes in brain structure and function to those typically observed in psychiatric patients, notably in the neural connections between limbic and cortical regions [18,19,20,21,22]. These neural differences may serve as partially heritable intermediate processes that mediate the link from genetic and environmental risk to more complex mental health symptamotology [23,24,25].
While the classification of psychiatric disorders has historically segmented mental health conditions into distinct categories, recent genetic studies have challenged the assumptions of disorder-specific risk by identifying a large number of common genetic variants with small effects, many of which predispose to multiple mental disorders [26,27,28]. Growing evidence supports a high degree of genetic correlation across different conditions [29, 30], indicating that the etiological and phenotypic boundaries between psychiatric disorders may not be as clear-cut as previously thought. Emerging evidence also suggests that correlations between childhood maltreatment and mental illness are driven partly by genetic factors [31]. However, much remains unknown about how these genetic risks manifest early in development. A key question motivating this work is whether genetic signals associated with psychiatric conditions in adulthood—such as schizophrenia, depression, or psychosis—are already detectable in childhood, and if so, how they relate to emerging symptoms and brain function. While many psychiatric disorders do not typically present until adolescence or later [32], their underlying genetic liability may influence preceding behavioural or neurobiological patterns. Investigating these early correlates can offer insight into how genetic and environmental risk factors unfold across development.
This study investigates how genetic and environmental risk factors converge to influence the neural substrates of mental illness in childhood, using data from a large cohort of 9–10-year-olds drawn from the Adolescent Brain and Cognitive Development (ABCD) study. We address three key questions: (1) To what extent do genetic and phenotypic risks overlap across psychiatric conditions? (2) How do genetic and environmental factors shape behavioural and neural profiles in childhood? (3) And finally, to what extent do these dimensions of risk converge at the level of brain connectivity?
First, we estimate polygenic risk scores (PRS) for four mental health phenotypes: ADHD, Anxiety, Depression, and Psychosis. We then use Canonical Correlation Analysis (CCA) to assess the degree of shared genetic liability across these mental health conditions, uncovering overlapping dimensions of genetic risk that transcend traditional phenotypic boundaries. Next, we examine how these dimensions of genetic liability relate to history of adversity. Finally, we use Partial Least Squares (PLS) to investigate the cortico-limbic signature of genetic and environment risk and identify neural markers of mental illness.
Methods
Participants
The Adolescent Brain Cognitive Development (ABCD) study is a longitudinal cohort that involves 21 data acquisition sites across the US and follows over 11,000 children aged 9–10 for 10 years into early adulthood [33, 34]. The study was designed to approximate the socio-demographic distribution of US children in this age group (Table S1). Participants were required to be aged 9–10 years at baseline for inclusion in the study. Those who lacked English language proficiency; suffered from severe sensory, intellectual, medical or neurological issues; or were unable to participate in MRI scanning were excluded. Recruitment details and data-collection procedures are described by Garavan et al. [33]. The current study uses data from participants with available genotype (n = 6535) and phenotype data as described below.
Mental health measures
The parent-reported Child Behaviour Checklist (CBCL) is made up of 113 items rated on a three-point scale (not true; somewhat or sometimes true; very often or always true; Achenbach, 2011). These items are then summed into several subscales. This study uses the three DSM-oriented scales from the CBCL that align with clinical disorder definitions: Attention-Deficit Hyperactivity Disorder (ADHD), Depressive Disorder, and Anxiety Disorder. The scales have good inter-interviewer and test-retest reliability [35]. Raw scores from the baseline assessment (T1), which capture the number of total questions endorsed, were used (range: 0–20). Additionally, the Prodromal Psychosis Scale-Brief Child Version (PPS) was used to measure psychotic symptoms [36]. The original screening questionnaire, developed for adolescents and adults, was modified for use with children [37, 38]. The raw score from T1, based on the number of total questions endorsed, was used (range: 0–117).
Early life adversity
A total of 24 questions were used to assess whether a child had been exposed to an adverse experience before the age of 10 (Table S2). Participants missing more than 15% of data on the adversity measures were removed from the analysis (n = 314) and the remaining missing answers were coded as ‘0’ (i.e., adversity not endorsed). These responses were coded as 0 because sensitivity analyses (reported in Supplement 1.2) revealed that either coding the missing responses as 1 (endorsing adversity) or using imputation resulted in estimates of adversity that were significantly higher than population prevalence estimates [39,40,41], indicating both approaches were heavily biased.
MRI acquisition and preprocessing
ABCD standard imaging protocols for resting-state functional MRI (rsfMRI) including acquisition, processing, and quality assurance procedures have been described in detail elsewhere [42, 43]. Briefly, the preprocessing pipeline includes within- and between-scan head-motion correction, distortion corrections, removal of initial frames, normalisation, demeaning, regression, and temporal filtering [44, 45]. Average time courses for each region of interest (ROI) were calculated using FreeSurfer’s automated brain segmentation (aseg) and resampled to align with voxels from the fMRI data. Motion time courses are adjusted to account for signals linked to respiration (Hagler et al. [43]). This study uses imaging data from a subset of participants with 10 min of valid rsfMRI data below a framewise displacement threshold of 0.2 mm collected at T1 (n = 5995).
Using the processed rsfMRI data, parcellated time series were computed using a seed-based correlational approach [46]. Regions of interest (ROIs) were defined using the functional Gordon atlas template which comprises 352 ROIs (333 cortical) belonging to one of 13 networks (12 cortical and 1 subcortical) [47]. The functional connectivity between any two ROIs was estimated by calculating the lag-zero Pearson correlation coefficient of parcellated time series. This produced an ROI x ROI correlation matrix for each participant, which underwent an additional variance stabilization procedure using a Fischer z-transform [48].
Polygenic risk score calculation
Saliva and blood samples were collected from participants as part of the ABCD biospecimen collection [49]. DNA extraction, basic biospecimen quality control, and genotyping using the Affymetrix Axiom Smokescreen Array were performed at the Rutgers University Cell and DNA Repository. Genotypes were called from the raw intensities using the Affymetrix Power Tools and the Affymetrix Best Practice Workflow in batch processing. The genomic coordinates were aligned with genome build hg19. To ensure data quality, cohort-level quality control (QC) procedures were implemented, which are described in detail elsewhere [9]. Samples missing more than 20% on genotype calls and variants with more than 10% missing rates were excluded during the initial QC. Samples with excessive relatedness were also removed. Individuals with sex discrepancies, such as those with ambiguous gender calls, were removed. Missing genotype data was inputted using the TOPMED reference panel with genome build GRCh38 and aligned with genome build hg19 [50].
After obtaining the minimally QCed genomic data, individuals from axiom plate 461, which was identified as problematic, were removed as recommended by the ABCD guidelines. Cryptic relatedness among samples was assessed, and one member of each first- and second-degree related pair was excluded to avoid confounding effects. Ambiguous SNPs and those with a missing allele were removed. PLINK 2.0 [51] was used for QC steps. Genetic variants with a call rate below 90% and minor allele frequency (MAF) less than 0.01 were removed to retain only common variants. Variants showing a significant departure from Hardy-Weinberg equilibrium (p < 1e-6) were filtered out to mitigate potential genotyping errors. Genetically related individuals with a first or second degree relative were removed. Pruning was performed to remove any highly correlated SNPs. Principal component analysis (PCA) was performed on the genotyping data to identify genetically inferred population ancestries to be used as covariates in the PRS calculation. Individuals with a heterozygosity rate of F>3sd from the population mean were removed. Duplicate SNPs and samples with a mislabelled sex chromosome were removed. A total of 6535 individuals passed all filters and QC steps.
Polygenic risk scores (PRS) were calculated for each of the four mental health phenotypes (see ‘Mental health measures’ above) using PRSice-2 software [52] based on a set of selected variants selected from the following case-control GWAS studies: (1) ADHD [53]; (2) Anxiety [54]; (3) Depression [55]; (4) Schizophrenia (for psychotic symptoms) [56]. Each of the studies defined cases based on clinical criteria, and controls were selected from individuals without the respective mental disorder. Variants reaching genome-wide significance (p < 5e-8) and those showing suggestive associations (p < 1e-5) were included in the PRS calculation[57]. PRSice-2 was run with the recommended settings, such as clumping and LD pruning thresholds, to control for linkage disequilibrium (LD) between selected variants [52]. Effect sizes and their standard errors for the selected variants were extracted from the GWAS summary statistics from each of the four studies. The PRS scores were calculated by summing the products of the effect sizes and the number of risk alleles carried by each individual at the selected variants for each of the four mental health phenotypes. The PRS calculation was also repeated separately for individuals from genetically inferred European (n = 5679) and non-European (n = 865) ancestries for sensitivity analyses. Descriptive statistics for study variables are shown in Table S40.
Statistical analysis
Identifying shared dimensions of genetic risk
Canonical Correlation Analysis (CCA) was performed using the ‘CCA’ package in R [58] to explore the underlying relationships between the four PRS scores (Set A) and their respective phenotypes (Set B) and to identify patterns of correlation across the two datasets. We applied Canonical Correlation Analysis to identify latent overlapping factors between the set of polygenic risk scores and behavioural measures of mental health. This analysis yields paired linear combinations (canonical variates) of variables that are maximally correlated with each other [59]. Thus, CCA provides the ability to assess the extent of shared genetic variance across different mental health phenotypes. In contrast to other dimensionality-reduction techniques, CCA computes the components in the predictors and outcomes simultaneously such that they are maximally correlated with each other [60].
We calculated PRS component scores by adding up individual scores for each mental health dimension, weighted accordingly. Similarly, phenotype component scores were derived from a weighted sum of four mental health phenotypes. The first and second canonical correlations indicate the strength of associations between corresponding variate pairs, ranked by their correlation strength.
To determine the statistical significance of these correlations, we generated p-values using F-approximations of various test statistics. Canonical coefficients (or weights) were used to identify which variables had the strongest influence on each variate. We conducted a series of correlation analyses to assess how strongly each variable in our model was associated with component scores, providing a clearer interpretation than using canonical weights alone. As a sensitivity check for potential confounding due to genetic population differences, we performed supplementary CCA analyses. These included population substructure principal components (PCs) as covariates for the entire sample and specifically for individuals of European ancestry.
Finally, multiple linear regressions were conducted to assess how much variance in each mental health phenotype is explained by each of the PRS components, controlling age, sex, and the first 6 population substructure PCs. This allowed us to assess whether the PRS components explain more (or less) variance than disorder-specific PRS scores for each mental health phenotype. Because CCA variates are orthogonal to each other, meaning that each additional variate explains only the remaining variance not accounted for by previous components, all PRS component scores were entered jointly as predictors in the regression models. Confidence intervals for the multiple regressions were generated using bootstrapping with 1000 replicates.
Relationship between genetic liability for mental-ill health and adversity
To assess possible overlapping liability between genetic risk and adversity, multiple linear regressions were performed to examine the unique contribution of each PRS component score to variation in cumulative adversity, controlling for the respective phenotype component score, age, sex, and the first 6 population substructure principle components (PCs). Genetic population PCs were included as covariates to mitigate spurious associations between ancestry-based population-level stratification and outcomes of interest. For instance, studies have demonstrated correlations between variation in population structure and certain brain imaging features, as well as the prevalence of poverty in the US [61, 62]. Failure to control for genetically-defined population structure can therefore in spurious associations between brain measures, mental health, and adversity driven by uncorrected for population stratification within a given sample.
Next, multiple hierarchical linear regressions were conducted to assess whether PRS and adversity both independently predict mental health phenotypes and whether there is an interaction effect between genetic risk and the environment. Cumulative adversity scores were entered alongside each PRS component as predictors for the respective phenotype component, controlling age, sex, and the first 6 population substructure PCs. Interaction terms between PRS variates and adversity were added to the models as a second step. Change in R squared between the steps was used to assess differences in the overall model after adding the interaction term with corresponding significance tests.
Cortico-limbic signature of genetic and environmental risk
In our final analyses, we leveraged partial least squares (PLS) to assess how genetic and environmental risk covaries with cortico-limbic global and regional connectivity. PLS is a data-reduction technique well suited to capturing covariance and explaining complex relationships between a large set of noisy or multicollinear variables. It models the relationship between predictor and outcome variables by simultaneously projecting them to a new space to obtain a set of orthogonal latent variables that represent linear combinations of the original variables [63, 64]. We conducted two separate PLS analyses to model the covariance between adversity/PRS and (1) cortico-limbic global connectivity; and (2) cortico-limbic regional connectivity (regional-PLS).
In the first PLS, we modelled the four mental health PRSs and cumulative adversity as joint predictor variables (joint model), and connectivity between the limbic network and each of the 12 cortical networks as the outcome. We then repeated this using only the PRS scores (PRS-only model) and then only cumulative adversity (adversity-only model) as the predictor variables to assess unique associations between different dimensions of risk and cortico-limbic network connectivity. In our second PLS, we modelled the joint, PRS-only, and adversity-only predictors with connectivity between specific limbic regions (n = 19) and each of the cortical networks as the outcome to assess possible regional heterogeneity within the limbic network.
The pcorr significance values for the PLS components were obtained by permuting the data 10,000 times and comparing the observed coefficients relative to their null distributions. Variable importance projection (VIP) was used to assess the relative importance of each variable in the model, with scores above 1 considered most influential in terms of their explanatory power. The stability of item loadings was assessed by averaging the mean squared error of prediction, R2, and Q2 across 10 cross-validation runs (nrepeat=10, folds=10), with a significance threshold of 0.01 for improvement in component error rate. To assess the stability of the model loadings, we employed a non-parametric bootstrapping approach with 1000 resampling iterations to obtain standard error, t-values, confidence intervals and p-values for each item. Each bootstrap sample was created by randomly resampling with replacement from the original dataset ensuring that the matrix structure for both predictor and response variables was preserved. Variables were scaled to unit variance prior to analysis. We regressed the latent variables obtained from each PLS model to obtain the latent component slope, with age, sex, and the 6 PCs added as covariates. Mental health phenotype scores were added as a second set of covariates. In the PRS-only model, adversity was added as a final covariate. In the adversity-only model, PRS scores were added as final covariates. For any significant regression results, we tested for possible mediating effects of the component response scores (i.e., latent scores based on cortico-limbic connectivity measures) on the association between the component predictor scores (i.e., latent scores based on PRS/adversity measures) and mental health. One thousand bootstrap samples were used to estimate the 95% confidence intervals for indirect effects using the bias-corrected percentile method proposed by Biesanz and colleagues (2010). Analyses were performed in R using the MixOmics [65], caret [66] and mdatools [67].
Results
Polygenic risk scores
GWAS summary statistics from four mental health disorders (ADHD; Anxiety; Depression; Psychosis) were used to calculate PRS scores for four mental health phenotypes in the ABCD cohort for 6535 participants who passed all filters and QC steps (Figures S1–S6; Table S3). Hierarchical linear regressions assessing how much variance in each mental health phenotype is explained by each of the PRS scores, with age, sex, and the first 6 population substructure PCs as covariates are reported in Tables S4–S7. All four PRS scores were significantly correlated with their respective mental health phenotype and with adversity (Fig. 1a). The polygenic risk scores accounted for less than 1% of variance in mental health phenotypes, a pattern consistent with prior large-scale studies showing that PRS typically explain between 0.1–3% of variance in behavior, even in adult populations [68, 69].
a Correlations between genotype scores, their respective phenotypes, and adversity. b Scaled canonical correlation coefficients for each canonical variate. r= canonical correlation between each set of canonical components. c Canonical loadings showing the strength and significance of correlation of all variables with each canonical component. d Comparison of variance explained by disorder-specific PRS scores and the two PRS components. prsCVgeneral= PRS component scores from the first canonical variate representing general psychopathology. phenoCVgeneral= Phenotype component scores from the first canonical variate representing general psychopathology. prsCVneurodev= PRS component scores from the second canonical variate representing neurodevelopmental-specific variance. phenoCVneurodev= Phenotype component scores from the second canonical variate representing neurodevelopmental-specific variance. *p < 0.05, **p < 0.01, ***p < 0.001.
Shared dimensions of genetic risk
Canonical Correlation Analysis (CCA) was used to explore the underlying relationships between the four PRS scores and their respective phenotypes and identify possible genetic and phenotypic overlap across the four mental health conditions. The CCA yielded two significant canonical variates of modest strength with the following canonical correlation coefficients: Canonical Variate 1 (r = 0.15; Wilks’ λ = 0.97, F (16, 19950.12) = 13.23, p < 0.001); Canonical Variate 2 (r = 0.09; Wilks’ λ = 0.99, F (9, 15894.89) = 6.43, p < 0.001; Figure S7). The canonical loadings showing the corresponding strength and significance of correlations between disorder-specific mental health variables and each of the obtained genetic and phenotypic component scores are shown in Fig. 1 and Table S8. These loadings suggest that the first canonical variate represents a general risk for psychopathology across multiple conditions (henceforth referred to as ‘CVgeneral’), whereas the second represents unique neurodevelopmental-specific variance, characterised by positive loadings only from prsADHD (henceforth referred to as ‘CVneurodev’).
Multiple linear regressions demonstrating the amount variance in each mental health phenotype explained by the two PRS components (prsCVgeneral & prsCVneurodev), controlling for age, sex, and 6 genetically inferred population ancestry principal components (PCs) and comparing these results to the predictive value of the disorder-specific PRS scores are shown in Fig. 1 (also Tables S9–S12). The results did not qualitatively change when including population substructure PCs as covariates for the entire sample (Tables S34–S36) and for individuals with European ancestry only (Tables S37–S39). Overall, this suggests that genetic liability for different mental health conditions can be reduced to two independent transdiagnostic dimensions to predict variance in the respective behavioural phenotypes.
Relationship between genetic liability for mental-ill health and adversity
To investigate whether genetic liability for mental illness is related to adversity, multiple linear regressions were conducted with prsCVgeneral and prsCVneurodev as predictors, controlling for the respective CVpheno scores, age, sex, and the 6 PCs. prsCVgeneral (b = 0.089, p < 0.001), but not prsCVneurodev (b = 0.000, p = 0.99), was positively related to adversity (Tables S13–S14). prsCVgeneral (b = 0.116, p < 0.001) and adversity (b = 0.243, p < 0.001) both independently predicted phenoCVgeneral, but there was no interaction effect (Fig. 2a, c; full results in Table S19). In contrast, prsCVneurodev (b = 0.084, p < 0.001) and adversity (b = −0.098, p < 0.001) both independently predicted phenoCVneurodev— with similar strength but with opposing directions of effect— and there was also a significant interaction between prsCVneurodev and adversity (b = 0.043, R² change = 0.001, p = 0.006), indicating that increases in the effect of either variable attenuate the influence of the other (Fig. 2b, d; Table S20).
a, b Association between genetic liability components and adversity. prsCVgeneral= PRS component scores from the first canonical variate representing general psychopathology. prsCVneurodev= PRS component scores from the first canonical variate representing neurodevelopmental-specific variance. Regression models include age, sex, the first 6 population components (PCs), and the respective phenotype measures as covariates. c, d Prediction of phenotype components by PRS components, adversity, and their interaction. Regressions model the predictive value for each phenotype component separately, including age, sex, and the first 6 population components (PCs) as covariates. ***p < 0.001.
To test whether this pattern was specific to particular types of adversity, we repeated these analyses using four disaggregated adversity dimensions (household/community instability, physical/sexual abuse, parental neglect, financial difficulties) identified in prior work [70]. In the prsCVgeneral models, interaction effects were of similar magnitude across multiple adversity domains. In the prsCVneurodev model, the interaction effect was significant for the household/community instability factor, with a similar magnitude to the cumulative risk score (Tables S1–S2). This suggests that the composite adversity index offers a stable and efficient way to capture shared variance across adversity dimensions that is relevant for gene–environment interplay. The additional exploration of whether the effect holds across more specific adversity domains reveals some nuance to the prsCVneurodev model, with household/community instability being the strongest driver of that interaction.
Cortico-limbic signature of genetic and environmental risk
Cortico-limbic global connectivity
The first PLS was used to model the covariance between genetic/environmental risk and connectivity between the entire limbic network (n = 1) and all cortical networks (n = 12). When using the four PRSs and adversity as joint predictors (joint model), a single significant component emerged (pperm < 0.001), explaining 33% of the variance in the predictors and 43% of the variance in the cortico-limbic network connections (Table S25). The correlation between the latent components was b = 0.03, SE = 0.009, p < 0.001 when controlling for age, sex and the 6PCs, and remained significant when mental health phenotype scores were added as covariates (b = 0.03, SE = 0.009, p < 0.001).
When using only the four PRSs as predictor variables (PRS-only model), a single significant component emerged (pperm < 0.001), explaining 42% of the variance in the PRSs and 43% of the variance in the cortico-limbic network connections (Fig. 3; Table S26). The correlation between the latent components was b = 0.07, SE= 0.034, p = 0.04 when controlling for age, sex and the 6PCs, and remained significant when mental health phenotype scores were added as covariates (b = 0.07, SE = 0.034, p = 0.05). However, the CCA model became non-significant when adversity was added as a covariate (b = 0.05, SE = 0.034, p = 0.11).
PLS loadings of cortico-limbic network-level connectivity on the PRS-only model and adversity-only model. Only significant loadings with a VIP score above 1 shown in figure. AD auditory; CO cingulo-opercular; CP cingulo-parietal; DA dorsal attention; DF default; FP frontoparietal; RST retrosplenial temporal; SA salience; SMH sensorimotor hand; SMM sensorimotor mouth; VA ventral attention; VS visual.
When using only cumulative adversity as the predictor (adversity-only model), a single significant component emerged (pperm < 0.001), explaining 100% of the variance in adversity and 42% of the variance in the cortico-limbic network connections (Fig. 3; Table S27). The correlation between the latent components was b = 0.03, SE = 0.008, p < 0.001 when controlling for age, sex and the 6PCs, and remained significant when mental health phenotypes scores (b = 0.03, SE = 0.007, p < 0.001) and the PRS scores (b = 0.03, SE = 0.007, p < 0.001) were added as covariates. The cortico-limbic network component significantly mediated the association between the adversity component and symptoms of depression and psychosis (p’s = 0.032–0.033; Table S28).
Together, these analyses suggest that while adversity and the four genetic liability factors map onto common reductions in cortico-limbic connectivity at the global level, adversity captures much of this shared variance. The mediation analyses underscore the role of cortico-limbic circuitry as a potential neural pathway through which these risk factors may influence mental health symptomatology.
Cortico-limbic regional connectivity
A second PLS was used to model the covariance between genetic/environmental risk and connectivity between individual limbic regions (n = 19) and cortical networks (n = 12). When using the PRSs and adversity as joint predictors (joint model), a single significant component emerged (pperm < 0.001), explaining 33% of the variance in the predictors and 10% of the variance in the cortico-limbic regional connections. There were 70 significant regional connection loadings out of a total of 228 (Table S29). The correlation between the latent components was b = 0.022, SE = 0.04, p < 0.001 when controlling for age, sex and the 6PCs, and remained significant when mental health phenotype scores were added as covariates (b = 0.021, SE = 0.004, p < 0.001).
When using only the PRSs as predictor variables (PRS-only model), a single significant component emerged (pperm < 0.001), explaining 42% of the variance in the PRSs and 10% of the variance in the cortico-limbic regional connections. There were 73 significant connections out of a total of 228 (Fig. 4; Table S30). The correlation between the latent components was b = 0.015, SE = 0.005, p < 0.001 when controlling for age, sex and the 6PCs, and remained significant when mental health phenotype scores (b = 0.014, SE = 0.005 p = 0.001) and adversity (b = 0.013, SE = 0.005, p = 0.005) were added as covariates. The cortico-limbic regional component significantly mediated the association between the PRS component and symptoms of psychosis (b = −0.021, p = 0.038; Table S31).
The cortico-limbic regional connections identified by the partial least squares (PLS) model. PRS (polygenic risk): connections that significantly loaded onto the PLS model for genetic liability. Adversity: Connections that significantly loaded onto the PLS model for adversity. PRS unique: Connections that are uniquely associated with genetic liability. Adversity unique: Connections that are uniquely associated with adversity. Only significant loadings with a VIP score above 1 shown in figure. AD auditory; CO cingulo-opercular; CP cingulo-parietal; DA dorsal attention; DF default; FP frontoparietal; RST retrosplenial temporal; SA salience; SMH sensorimotor hand; SMM sensorimotor mouth; VA ventral attention; VS visual. The PRS and adversity models differed on 41 out of 147 significant loadings.
When using only cumulative adversity as the predictor (adversity-only model), a single significant component emerged (pperm < 0.001), explaining 100% of the variance in the adversity and 10% of the variance in the cortico-limbic regional connections. There were 74 significant regional connections out of a total of 228 (Fig. 4; Table S32). The adversity-only model differed from the PRS-only model on 41 out of 147 significant loadings. Notably, the PRS-only model had greater loadings of limbic connectivity with the frontoparietal and visual networks, while the adversity-only model had greater loadings of limbic connectivity with the salience network. In contrast, there were few differences between the two models in terms of limbic connectivity with the cingulo-opercular, cingulo-parietal, and sensorimotor mouth networks. The correlation between the latent components was b = 0.023, SE = 0.004, p < 0.001 when controlling for age, sex and the 6PCs, and remained significant when mental health phenotypes scores (b = 0.023, SE = 0.004, p < 0.001) and the PRS scores (b = 0.021, SE = 0.004, p < 0.001) were added as covariates. The cortico-limbic regional component significantly mediated the association between the adversity component and symptoms of psychosis (b = −0.042, p = 0.006; Table S33).
Together, these analyses suggest that while adversity and the four genetic liability factors correspond to broadly similar regional differences in cortico-limbic connectivity, distinct patterns of connectivity associated with each dimension of risk become evident at the regional level of analysis. The mediation analyses underscore the role of cortico-limbic circuitry as a potential neural pathway through which both genetic and environmental risk factors may independently influence mental health symptomatology.
Discussion
We investigated relationships between polygenic risk for mental illness, childhood adversity, and cortico-limbic connectivity in a large developmental sample drawn from the ABCD study and uncovered two novel findings. First, genetic liability for ADHD, anxiety, depression, and psychosis can be reduced to two pleotropic dimensions of risk, each of which uniquely interact with exposure to adversity. Second, genetic and environmental risk factors overlap in their neural correlates. This shared neural signature indicates a gene-environment correlation at the level of the brain, with substantial implications for future research on the neural correlates and etiology of mental illness.
Our findings add to a growing body of evidence that traditional diagnostic boundaries may not map onto distinct neurobiological processes [27, 71, 72]. Given the common genetic origins and phenotypic overlap between disorder categories [28, 29, 73], existing disorder-specific GWAS studies may require re-evaluation. Furthermore, our study demonstrates significant correlations between genetic liability for psychopathology and environmental adversity in this sample [31, 74, 75]. Specifically, we found that genetic liability for general psychopathology was positively associated with adversity, suggesting an additive effect, whereby both factors appear to exert independent, cumulative influence on symptom severity [76]. In contrast, the association between adversity and neurodevelopmental symptoms was attenuated in individuals with higher genetic risk, suggesting a compensatory or interaction effect whereby one source of risk (genetic or environmental) reduces the influence of the other on symptom presentation. This points to the likelihood that neurodevelopmental symptoms may arise from combinations of genetic and environmental factors unique from other symptom domains. This could reflect a degree of etiological independence: genetic factors underlying ADHD/neurodevelopmental problems may influence brain development regardless of the child’s environmental adversity level. However, it should be noted that the adversity composite included items like parental psychiatric history and family psychosocial stressors. As such, higher polygenic risk for general psychopathology is more likely to co-occur with certain adversity indicators due to shared familial risk factors, potentially inflating the observed correlation for general liability.
Our study also highlights that environmental and genetic risk largely overlap in the neural connections that underlie mental health symptom variance. Both adversity and genetic liability were associated with reduced functional connectivity between the limbic and the cingulo-opercular, cingulo-parietal, visual, default, and sensorimotor networks. This global pattern of altered connectivity indicates that widespread disruptions in cortico-limbic connectivity are a core feature of transdiagnostic symptomatology [77,78,79,80]. In other words, pleotropic risk genes for multiple disorders and environmental risk factors increase susceptibility to a variety of clinically-distinct psychiatric conditions though non-specific changes in cortico-limbic connections by altering stress-susceptible systems in similar ways [15,16,17, 81].
However, our regional analyses also revealed notable heterogeneity within the limbic network, with 41 out of 147 cortico-limbic connections showing unique associations with genetic risk. The strongest effects emerged in connections with the frontoparietal and visual networks, suggesting that these circuits may be particularly sensitive to polygenic influences [82, 83]. These modest and spatially distributed PRS–brain associations are in line with findings from large-scale imaging genetics studies, which consistently report that individual genetic variants account for less than 0.1% of variance in brain structure or function, and that polygenic effects are typically diffuse rather than localized [83, 84]. Thus, although the observed genetic effects were weaker than those for adversity, their distribution and magnitude are consistent with current expectations for polygenic contributions to brain connectivity. Importantly, as GWAS sample sizes grow, PRS effect sizes tend to increase not by implicating entirely new biological mechanisms, but by capturing more variants within the same underlying molecular and cellular pathways [85, 86]. This suggests that even modest early signals may reflect biologically meaningful risk processes that become more apparent over time.
The finding that adversity accounted for the entirety of the genetic variance at the global level suggests that many of the previously identified neural markers of psychopathology may be capturing adversity-related variance, and vice-versa. In other words, the neural features stemming from early adversity are likely conflated with those that predispose individuals to mental illness in the existing literature [19]. Although many recent studies incorporate controls for co-occurring psychiatric symptoms, this has not been a consistent feature of earlier research on adversity-related neural outcomes [12, 19]. Even in contemporary work, approaches vary widely, with some studies adjusting for broad symptom indices, others focusing only on select diagnoses, and many omitting psychiatric covariates altogether [85, 87,88,89]. This methodological inconsistency complicates efforts to isolate the unique neurobiological effects of adversity and genetic risk and should be carefully considered when interpreting findings across the existing literature.
Efforts to identify genetically driven neural endophenotypes of mental illness [90] would do well to recognise this as a key methodological challenge for the field. Future research would benefit from stratifying mental health conditions based not only on clinical symptoms and neurobiological features, but also on genetic and environmental risk factors. Such an approach could pave the way towards precise diagnoses, endophenotypes, and tailored treatments [91,92,93] that are more effective for certain individuals and populations [94,95,96].
The strengths of this study lie in its application of diverse methods to capture multicollinear associations between genetic liability, environmental risk, and functional brain connectivity. However, some limitations must also be acknowledged. First, the study sample is representative of the US context and was restricted to children aged 9–10, limiting the generalisability of the findings to other demographic groups and age ranges. Future longitudinal research should endeavour to replicate these findings in diverse demographic groups across development. Second, while the behavioural expression of mental health difficulties may vary at different developmental time points [97, 98], this study only considered four mental health conditions due to data availability constraints. Third, while the use of dimensional symptom scales circumvents some shortcomings inherent to clinical diagnostic categories [99], it is worth noting that only a small fraction of the sample met the threshold for a clinical diagnosis. This is likely because our sample was composed of children aged 9–10, prior to the typical onset of psychiatric disorders like schizophrenia and major depression [32]. Consequently, polygenic risk for these conditions may not yet be phenotypically expressed in behavior or neurobiology [100]. Indeed, the stronger association of adversity with functional connectivity in our study may reflect the relatively low behavioural symptom variance explained by the PRS scores. Furthermore, connectivity differences related to psychopathology risk may still be emerging at age 9–10, given the ongoing maturation of cortico-limbic networks [101, 102]. While our findings suggest shared genetic and neural patterns across multiple symptom domains, we emphasize that these associations are modest in magnitude and developmentally constrained. At age 9–10, many psychiatric conditions have not yet fully emerged, and the neural signatures of risk may still be evolving. Accordingly, the associations we observed likely reflect early, subclinical markers of genetic liability, rather than predictors of imminent psychopathology. Nonetheless, these findings contribute to a growing transdiagnostic literature by demonstrating that polygenic and environmental risk factors can converge on shared neurobiological pathways prior to clinical onset [69, 103]. Longitudinal follow-up of this cohort will be necessary to determine whether these early associations strengthen or change over time [68]. It is also necessary to validate these findings using genetic methods better suited to capturing shared genetic signals across multiple conditions [104].
In line with established cumulative risk frameworks [105], we used a composite adversity score to capture the overall burden of early life adversity. Aggregating across diverse exposures helps reduce measurement error and reflects the fact that children rarely experience isolated adverse events; rather, multiple adversities often co-occur and interact developmentally [106]. While this approach maximizes power and supports dimensional modelling across neurobehavioral domains, we acknowledge that collapsing adversity into a single index may potentially mask distinct effects of specific adversity types. However, our supplemental analyses suggest that disaggregation did not reveal stronger, but did suggest that household/community instability to be the strongest driver with regard to the prsCVneurodev interaction. In prior work using the same ABCD dataset [70], we also found that attempts to disaggregate adversity into threat and deprivation dimensions were limited by multicollinearity and did not yield clearly distinct neural correlates. Nonetheless, future work could benefit from more refined measurement models in samples where these constructs are psychometrically separable, to explore potential specificity of effects.
In conclusion, this study demonstrates that comparable neural differences can emerge from both endogenous (genetic) and exogenous (environmental) risk factors. Our findings add to the growing evidence of overlapping etiological influences across symptom domains, even in a pediatric sample. Future longitudinal research will be critical to determine the clinical significance of these early patterns and to further elucidate how genetic and environmental factors jointly shape the neural substrates of mental illness over the course of development.
Data availability
The datasets analysed during the current study are available in the NIMH Data Archive (NDA) repository, https://doi.org/10.15154/1523041.
Code availability
The underlying code for this study is not publicly available but may be made available to qualified researchers on reasonable request from the corresponding author.
References
Wainberg, M, Jacobs, GR, Voineskos, AN & Tripathy, SJ Neurobiological, familial and genetic risk factors for dimensional psychopathology in the Adolescent Brain Cognitive Development study. Mol Psychiatry 1–11 (2022) https://doi.org/10.1038/s41380-022-01522-w.
Lynch CJ, Gunning FM, Liston C. Causes and consequences of diagnostic heterogeneity in depression: paths to discovering novel biological depression subtypes. Biol Psychiatry. 2020;88:83–94.
Choi KW, Wilson M, Ge T, Kandola A, Patel CJ, Lee SH, Smoller JW. Integrative analysis of genomic and exposomic influences on youth mental health. J Child Psychol Psychiatry. 2022;63:1196–205.
Kwong ASF, López-López JA, Hammerton G, Manley D, Timpson NJ, Leckie G, Pearson RM. Genetic and environmental risk factors associated with trajectories of depression symptoms from adolescence to young adulthood. JAMA Netw Open. 2019;2:e196587–e196587.
Mcgrath JJ, Mortensen PB, Visscher PM, Wray NR. Where GWAS and epidemiology meet: opportunities for the simultaneous study of genetic and environmental risk factors in schizophrenia. Schizophr Bull. 2013;39:955–9.
Hicks BM, South SC, DiRago AC, Iacono WG, McGue M. Environmental adversity increases genetic risk for externalizing disorders. Arch Gen Psychiatry. 2009;66:640.
Pagliaccio, D, Barch, DM Early life adversity and risk for depression: alterations in cortisol and brain structure and function as mediating mechanisms. Syst Neurosci Depress 29–77 (2016) https://doi.org/10.1016/B978-0-12-802456-0.00002-9.
Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13.
Fan CC, Loughnan R, Wilson S, Hewitt JK, Agrawal A, Dowling G, et al. Genotype data and derived genetic instruments of adolescent brain cognitive development study® for better understanding of human brain development. Behav Genet. 2023;53:159–68.
Mehta MA, Gore-Langton E, Golembo N, Colvert E, Williams SCR, et al. Hyporesponsive reward anticipation in the basal ganglia following severe institutional deprivation early in life. J Cogn Neurosci. 2010;22:2316–25.
Miller, JG, Chahal, R & Gotlib, IH Early Life Stress and Neurodevelopment in Adolescence: Implications for Risk and Adaptation. in Neuroscience of Social Stress (eds. Miczek, KA & Sinha, R) 313-39 (Springer, Berlin, Heidelberg, 2022). https://doi.org/10.1007/7854_2022_302.
Teicher MH, Samson JA. Annual Research Review: Enduring neurobiological effects of childhood abuse and neglect. J Child Psychol Psychiatry. 2016;57:241.
Tottenham N, Hare TA, Millner A, Gilhooly T, Zevin JD, Casey BJ. Elevated amygdala response to faces following early deprivation. Dev Sci. 2011;14:190–204.
Holz NE, Berhe O, Sacu S, Schwarz E, Tesarz J, Heim CM, et al. Early social adversity, altered brain functional connectivity, and mental health. Biol Psychiatry. 2023;93:430–41.
Colich NL, Rosen ML, Williams ES, McLaughlin KA. Biological aging in childhood and adolescence following experiences of threat and deprivation: a systematic review and meta-analysis. Psychol Bull. 2020;146:721.
McLaughlin KA, Sheridan MA, Tibu F, Fox NA, Zeanah CH, Nelson CA. Causal effects of the early caregiving environment on development of stress response systems in children. Proc Natl Acad Sci. 2015;112:5637–42.
Teicher MH, Khan A. Childhood maltreatment, cortical and amygdala morphometry, functional connectivity, laterality, and psychopathology. Child Maltreat. 2019;24:458–65.
Sheridan MA, Sarsour K, Jutte D, D’Esposito M, Boyce WT. The impact of social disparity on prefrontal function in childhood. PLoS One. 2012;7:e35744.
Teicher MH, Samson JA. Childhood maltreatment and psychopathology: a case for ecophenotypic variants as clinically and neurobiologically distinct subtypes. Am J Psychiatry. 2013;170:1114–33. https://doi.org/10.1176/appi.ajp.2013.12070957.
Brieant AE, Sisk LM, Gee DG. Associations among negative life events, changes in cortico-limbic connectivity, and psychopathology in the ABCD Study. Dev Cogn Neurosci. 2021;52:101022.
Gee DG, Gabard-Durnam LJ, Flannery J, Goff B, Humphreys KL, Telzer EH, et al. Early developmental emergence of human amygdala-prefrontal connectivity after maternal deprivation. Proc Natl Acad Sci USA. 2013;110:15638–43.
Guadagno A, Belliveau C, Mechawar N, Walker CD. Effects of early life stress on the developing basolateral amygdala-prefrontal cortex circuit: the emerging role of local inhibition and perineuronal nets. Front Hum Neurosci. 2021;15:669120.
Blokland GAM, De Zubicaray GI, McMahon KL, Wright MJ. Genetic and environmental influences on neuroimaging phenotypes: a meta-analytical perspective on twin imaging studies. Twin Res Hum Genet. 2012;15:351–71.
Cannon TD, Keller MC. Endophenotypes in the genetic analyses of mental disorders. Annu Rev Clin Psychol. 2006;2:267–90. https://doi.org/10.1146/annurev.clinpsy.2.022305.095232.
Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: The seven pillars of RDoC. BMC Med. 2013;11:126.
Smoller JW, Kendler KK, Craddock N, Lee PH, Neale BM, Nurnberger JN, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet (London, England). 2013;381:1371–9.
Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–94.
Wu Y, Cao H, Baranova A, Huang H, Li S, Cai L, et al. Multi-trait analysis for genome-wide association study of five psychiatric disorders. Transl Psychiatry. 2020;10:1–11.
Tylee DS, Lee YK, Wendt FR, Pathak GA, Levey DF, De Angelis F, et al. An atlas of genetic correlations and genetically informed associations linking psychiatric and immune-related phenotypes. JAMA Psychiatry. 2022;79:667–76.
Lee PH, Anttila V, Won H, Feng YCA, Rosenthal J, Zhu Z, Tucker-Drob EM, et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell. 2019;179:1469–1482.e11.
Warrier V, Kwong ASF, Luo M, Dalvie S, Croft J, Sallis HM, et al. Gene–environment correlations and causal effects of childhood maltreatment on physical and mental health: a genetically informed approach. The Lancet Psychiatry. 2021;8:373–86.
Solmi M, Radua J, Olivola M, Croce E, Soardo L, Salazar de Pablo G, et al. Age at onset of mental disorders worldwide: large-scale meta-analysis of 192 epidemiological studies. Mol Psychiatry. 2022;27:281–95. https://doi.org/10.1038/s41380-021-01161-7.
Garavan H, Bartsch H, Conway K, Decastro A, Goldstein RZ, Heeringa S, et al. Recruiting the ABCD sample: Design considerations and procedures. Dev Cogn Neurosci. 2018;32:16–22.
Achenbach, TM Child Behavior Checklist. in Encyclopedia of Clinical Neuropsychology 546-52 (Springer New York, 2011). https://doi.org/10.1007/978-0-387-79948-3_1529.
Achenbach TM, Rescorla LA. Manual for the ASEBA school age forms & profiles. ASEBA. 2001;99:135.
Loewy RL, Pearson R, Vinogradov S, Bearden CE, Cannon TD. Psychosis risk screening with the Prodromal Questionnaire–brief version (PQ-B). Schizophr Res. 2011;129:42–46.
Karcher NR, Barch DM, Avenevoli S, Savill M, Huber RS, Simon TJ, et al. Assessment of the prodromal questionnaire–brief child version for measurement of self-reported psychoticlike experiences in childhood. JAMA Psychiatry. 2018;75:853–61.
Berman IS, McLaughlin KA, Tottenham N, Godfrey K, Seeman T, Loucks E, et al. Measuring early life adversity: a dimensional approach. Dev Psychopathol. 2022;34:499–511.
Finkelhor D, Ormrod R, Turner H, Hamby SL. The victimization of children and youth: a comprehensive, national survey. Child Maltreat. 2005;10:5–25.
McLaughlin KA, Green JG, Gruber MJ, Sampson NA, Zaslavsky AM, Kessler RC. Childhood adversities and first onset of psychiatric disorders in a national sample of US adolescents. Arch Gen Psychiatry. 2012;69:1151–60.
Struck N, Krug A, Yuksel D, Stein F, Schmitt S, Meller T, et al. Childhood maltreatment and adult mental disorders - the prevalence of different types of maltreatment and associations with age of onset and severity of symptoms. Psychiatry Res. 2020;293:113398.
Casey BJ, Cannonier T, Conley MI, Cohen AO, Barch DM, Heitzeg MM, et al. The Adolescent Brain Cognitive Development (ABCD) study: imaging acquisition across 21 sites. Dev Cogn Neurosci. 2018;32:43–54.
Hagler DJ, Hatton SN, Cornejo MD, Makowski C, Fair DA, Dick AS, et al. Image processing and analysis methods for the Adolescent Brain Cognitive Development Study. Neuroimage. 2019;202:116091.
Holland D, Kuperman JM, Dale AM. Efficient correction of inhomogeneous static magnetic field-induced distortion in Echo Planar Imaging. Neuroimage. 2010;50:175–83.
Jovicich J, Czanner S, Greve D, Haley E, Van Der Kouwe A, Gollub R, et al. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–43.
Van Dijk KRA, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol. 2010;103:297–321.
Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb Cortex. 2016;26:288–303.
Feczko, E, Earl, E, Perrone, A, Fair, D ABCD-BIDS Community Collection (ABCC). (2020) https://doi.org/10.17605/OSF.IO/PSV5M.
Uban KA, Horton MK, Jacobus J, Heyser C, Thompson WK, Tapert SF, et al. Biospecimens and the ABCD study: Rationale, methods of collection, measurement and early data. Dev Cogn Neurosci. 2018;32:97–106.
Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–9.
Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Choi SW, O’Reilly PF. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience. 2019;8:1–6.
Demontis D, Walters GB, Athanasiadis G, Walters R, Therrien K, Nielsen TT, et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat Genet. 2023;55:198–208.
Meier SM, Trontti K, Purves KL, Als TD, Grove J, Laine M, et al. Genetic variants associated with anxiety and stress-related disorders: a genome-wide association study and mouse-model study. JAMA Psychiatry. 2019;76:924–32.
Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81.
Richards AL, Cardno A, Harold G, Craddock NJ, Di Florio A, Jones L, et al. Genetic liabilities differentiating bipolar disorder, schizophrenia, and major depressive disorder, and phenotypic heterogeneity in bipolar disorder. JAMA Psychiatry. 2022;79:1032–9.
Marees AT, de Kluiver H, Stringer S, Vorspan F, Curis E, Marie-Claire C, et al. A tutorial on conducting genome‐wide association studies: Quality control and statistical analysis. Int J Methods Psychiatr Res. 2018;27:e1608.
González I, Déjean S, Martin PGP, Baccini A. CCA: an R package to extend canonical correlation analysis. J Stat Softw. 2008;23:1–14.
Wickramasinghe ND. Canonical correlation analysis: an introduction to a multivariate statistical analysis. J Coll Community Physicians Sri Lanka. 2019;25:37.
Hardoon DR, Szedmak S, Shawe-Taylor J. Canonical correlation analysis: An overview with application to learning methods. Neural Comput. 2004;16:2639–64.
Huang, TH, Loughnan, R, Thompson, WK, Fan, CC The impact of population stratification on the analysis of multimodal neuroimaging derived measures. bioRxiv: 2022.08.06.503037 [Preprint]. 2022. https://doi.org/10.1101/2022.08.06.503037.
Fan CC, Loughnan R, Wilson S, Hewitt JK, Agrawal A, Dowling G, et al. Modeling the 3D geometry of the cortical surface with genetic ancestry. Curr Biol. 2015;25:1988–92.
Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst. 2001;58:109–30.
Biesanz JC, Falk CF, Savalei V. Assessing mediational models: testing and interval estimation for indirect effects. Multivariate Behav Res. 2010;45:661–701. https://doi.org/10.1080/00273171.2010.498292.
Rohart F, Gautier B, Singh A, Lê Cao KA. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLOS Comput Biol. 2017;13:e1005752.
Kuhn, M Classification and Regression Training. (2022).
Kucheryavskiy S. mdatools – R package for chemometrics. Chemom Intell Lab Syst. 2020;198:103937.
Allegrini AG, Cheesman R, Rimfeld K, Selzam S, Pingault JB, Eley TC, Plomin R. The p factor: genetic analyses support a general dimension of psychopathology in childhood and adolescence. J Child Psychol Psychiatry Allied Discipl. 2020;61:30–39. https://doi.org/10.1111/jcpp.13113.
Elliott ML, Romer A, Knodt AR, Hariri AR. A connectome-wide functional signature of transdiagnostic risk for mental illness. Biological Psychiatry. 2018;84:452–9. https://doi.org/10.1016/j.biopsych.2018.03.012.
Vedechkina M, Astle DE, Holmes J. Dimensions of early life adversity and their associations with functional brain organisation. Imaging Neuroscience. 2024;2:145.
Goodkind M, Eickhoff SB, Oathes DJ, Jiang Y, Chang A, Jones-Hagata LB, et al. Identification of a common neurobiological substrate for mental illness. JAMA Psychiatry. 2015;72:305–15.
McTeague LM, Huemer J, Carreon DM, Jiang Y, Eickhoff SB, Etkin A. Identification of common neural circuit disruptions in cognitive control across psychiatric disorders. Am J Psychiatry. 2017;174:676–85.
Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757.
Richardson TG, Harrison S, Hemani G, Smith GD. An atlas of polygenic risk score associations to highlight putative causal relationships across the human phenome. Elife. 2019;8:e43657.
Leppert B, Millard LAC, Riglin L, Smith GD, Thapar A, Tilling K, et al. A cross-disorder PRS-pheWAS of 5 major psychiatric disorders in UK Biobank. PLOS Genet. 2020;16:e1008185.
Coleman JRI, Peyrot WJ, Purves KL, Davis KAS, Rayner C, Choi SW, et al. Genome-wide gene-environment analyses of major depressive disorder and reported lifetime traumatic experiences in UK Biobank. Mol Psychiatry. 2020;25:1430–46.
Bassett DS, Xia CH, Satterthwaite TD. Understanding the emergence of neuropsychiatric disorders with network neuroscience. Biol Psychiatry Cogn Neurosci Neuroimaging. 2018;3:742–53.
Cao M, Huang H, Peng Y, Dong Q, He Y. Toward developmental connectomics of the human brain. Front Neuroanat. 2016;10:25.
Satterthwaite TD, Baker JT. How can studies of resting-state functional connectivity help us understand psychosis as a disorder of brain development? Curr Opin Neurobiol. 2015;0:85.
Bassett DS, Bullmore ET. Small-World Brain Networks Revisited. Neuroscientist. 2017;23:499–516.
Buckholtz JW, Meyer-Lindenberg A. Psychopathology and the human connectome: toward a transdiagnostic model of risk for mental illness. Neuron. 2012;74:990–1004.
Westwater ML, Mallard TT, Warrier V, Bethlehem RAI, Scheinost D, Grillon, et al. Assessing a multivariate model of brain-mediated genetic influences on disordered eating in the ABCD cohort. Nat Ment Heal. 2023;1:573–85.
Zhao B, Li T, Smith SM, Xiong D, Wang X, Yang Y, et al. Common variants contribute to intrinsic human brain functional networks. Nat Genet. 2022;54:508–17.
Smith SM, Douaud G, Chen W, Hanayik T, Alfaro-Almagro F, Sharp K, et al. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat Neurosci. 2021;24:737–45. https://doi.org/10.1038/s41593-021-00826-4.
Wray NR, Lin T, Austin J, McGrath JJ, Hickie IB, Murray GK, et al. From basic science to clinical application of polygenic risk scores: a primer. JAMA Psychiatry. 2021;78(1):101–9. https://doi.org/10.1001/jamapsychiatry.2020.304.
Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–86. https://doi.org/10.1016/j.cell.2017.05.038.
Chan SY, Ngoh ZM, Ong ZY, Teh AL, Lee MZ, Zhou JH, et al. The influence of early-life adversity on the coupling of structural and functional brain connectivity across childhood. Nat Mental Health. 2024;2:52–62. https://doi.org/10.1038/s44220-023-00162-5.
Silvers JA, Lumian DS, Gabard-Durnam L, Gee DG, Goff B, Fareri DS, et al. Previous institutionalization is followed by broader Amygdala-Hippocampal-PFC network connectivity during aversive learning in human development. J Neurosci. 2016;36:6420–30. https://doi.org/10.1523/JNEUROSCI.0038-16.2016.
Stinson EA, Sullivan RM, Navarro GY, Wallace AL, Larson CL, Lisdahl KM. Childhood adversity is associated with reduced BOLD response in inhibitory control regions amongst preadolescents from the ABCD study. Dev Cogn Neurosci. 2024;67:101378 https://doi.org/10.1016/j.dcn.2024.101378.
Prathikanti S, Weinberger DR. Psychiatric genetics–the new era: genetic research and some clinical implications. Br Med Bull. 2005;73–74:107–22.
Kanes S, Colquhoun H, Gunduz-Bruce H, Raines S, Arnold R, Schacterle A, et al. Brexanolone (SAGE-547 injection) in post-partum depression: a randomised controlled trial. Lancet. 2017;390:480–9.
Mayberg HS, Lozano AM, Voon V, McNeely HE, Seminowicz D, Hamani C, et al. Deep brain stimulation for treatment-resistant depression. Neuron. 2005;45:651–60.
Zarate CA, Singh JB, Carlson PJ, Brutsche NE, Ameli R, Luckenbaugh DA, et al. A randomized trial of an N-methyl-D-aspartate antagonist in treatment-resistant major depression. Arch Gen Psychiatry. 2006;63:856–64.
Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, et al. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med. 2016;23:28–38.
Van Nierop M, Viechtbauer W, Gunther N, Van Zelst C, De Graaf R, Ten Have M, et al. Childhood trauma is associated with a specific admixture of affective, anxiety, and psychosis symptoms cutting across traditional diagnostic boundaries. Psychol Med. 2015;45:1277–88.
van Nierop M, Bak M, de Graaf R, ten Have M, van Dorsselaer S, Bruggeman R, et al. The functional and clinical relevance of childhood trauma-related admixture of affective, anxious and psychosis symptoms. Acta Psychiatr Scand. 2016;133:91–101.
Maciejewski DF, van Lier PAC, Branje SJT, Meeus WHJ, Koot HM. A daily diary study on adolescent emotional experiences: measurement invariance and developmental trajectories. Psychol Assess. 2017;29:35–49.
Ochoa S, Usall J, Cobo J, Labad X, Kulkarni J. Gender differences in schizophrenia and first-episode psychosis: a comprehensive literature review. Schizophr Res Treatment. 2012;2012:1–9.
Astle DE, Holmes J, Kievit R, Gathercole SE. Annual Research Review: The transdiagnostic revolution in neurodevelopmental disorders. J Child Psychol Psychiatry. 2022;63:397–417.
Plomin R, DeFries JC, Knopik VS, Neiderhiser JM. Top 10 replicated findings from behavioral genetics. Perspect Psychol Sci. 2016;11:3–23.
Fair DA, Cohen AL, Power JD, Dosenbach NUF, Church JA, Miezin FM, et al. Functional brain networks develop from a “Local to Distributed” organization. PLOS Comput Biol. 2009;5:e1000381.
Khundrakpam BS, Reid A, Brauer J, Carbonell F, Lewis J, Ameis S, et al. Developmental changes in organization of structural brain networks. Cereb Cortex. 2013;23:2072–85.
Riglin L, Thapar AK, Leppert B, Martin J, Richards A, Anney R, et al. Using genetics to examine a general liability to childhood psychopathology. Behav Genet. 2020;50:213–20. https://doi.org/10.1007/s10519-019-09985-4.
Grotzinger AD, Mallard TT, Akingbuwa WA, Ip HF, Adams MJ, Lewis CM, et al. Genetic architecture of 11 major psychiatric disorders at biobehavioral, functional genomic and molecular genetic levels of analysis. Nat Genet. 2022;54:548–59.
Evans GW, Li D, Whipple SS. Cumulative risk and child development. Psychol Bull. 2013;139:1342–96. https://doi.org/10.1037/a0031808.
Smith KE, Pollak SD. Rethinking concepts and categories for understanding the neurodevelopmental effects of childhood adversity. Perspect Psychol Sci. 2021;16:67–93. https://doi.org/10.1177/1745691620920725.
Acknowledgements
This study was funded by the UK Medical Research Council, Grant MC-A0606-5PQ41. DA is supported by the Gnodde Goldman Sachs endowed Professorship in Neuroinformatics, The James S. McDonnell Foundation Opportunity Awards, and by the Templeton World Charity Foundation, Inc. (funder DOI 501100011730) under grant TWCF-2022-30510. All research at the Department of Psychiatry at the University of Cambridge is supported by the National Institute for Health and Care Research Cambridge Biomedical Research Centre (NIHR203312) and the NIHR Applied Research Collaboration East England. The funders played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript. Data used in the preparation of this article were obtained from the Adolescent Brain Cognitive Development (ABCD) Study (https://abcdstudy.org), held in the NIMH Data Archive (NDA). ABCD consortium investigators designed and implemented the study and/or provided data but did not participate in the analysis or writing of this report. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or ABCD consortium investigators. The ABCD Study is supported by the National Institutes of Health and additional federal partners under award numbers U01DA041048, U01DA050989, U01DA051016, U01DA041022, U01DA051018, U01DA051037, U01DA050987, U01DA041174, U01DA041106, U01DA041117, U01DA041028, U01DA041134, U01DA050988, U01DA051039, U01DA041156, U01DA041025, U01DA041120, U01DA051038, U01DA041148, U01DA041093, U01DA041089, U24DA041123, U24DA041147. A full list of supporters is available at https://abcdstudy.org/federal-partners.html.
Author information
Authors and Affiliations
Contributions
MV carried out the conceptualisation, methodology, data curation, investigation, formal analysis, visualisation, reviewing, writing and editing; JH carried out the conceptualisation, methodology, reviewing, writing, editing, and supervision; DA carried out the conceptualisation, methodology, reviewing, writing, editing, and supervision; VW carried out the methodology and reviewing. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations. The current study involves secondary analysis of data from the Adolescent Brain Cognitive Development (ABCD) Study. The ABCD Study obtained ethics approval from a central Institutional Review Board (University of California, San Diego) and site-specific ethics boards. Written informed consent and assent were obtained from parents/guardians and participants, respectively. The ABCD data accessed and analyzed in this study are fully anonymized and publicly available, and their use for secondary analysis was reviewed and approved under the original ABCD ethics protocols.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vedechkina, M., Holmes, J., Warrier, V. et al. A common neural signature between genetic and environmental risk for mental illness. Transl Psychiatry 15, 305 (2025). https://doi.org/10.1038/s41398-025-03513-1
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41398-025-03513-1