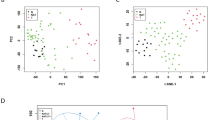
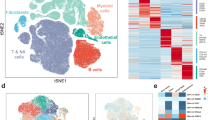
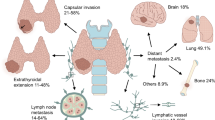
Abstract
Papillary thyroid cancer (PTC) is the most common endocrine cancer, with a good prognosis in most cases. However, aggressive PTC can metastasise or reoccur and become refractory disease. Therefore, it’s urgent to uncover new biomarkers for aggressive PTC. Accumulating evidence suggests that aberrant enhancers and targeted gene transcription drive the progression of PTC. To identify the cancer-specific enhancers and their downstream genes in PTC, we profiled the transcriptomes (RNA-seq) and enhancer-based epigenomic reorganisation (ChIP-seq) of cancer tissues and matched normal tissues from three PTC patients. Importantly, six candidate genes (RHBDF1, FAM20C, PHLDA2, TMPRSS6, LAD1, and BGN) were identified to be consistently upregulated by enhancers in PTC and correlated with prognosis. Further experiments verified the function of enhancers governing FAM20C in regulating PTC tumorigenesis, thereby unveiling a FAM20C-governed oncogenic mechanism for suppressing two cytokines (TNF-α and TGF-β) in PTC. Additionally, we demonstrated that a FAM20C inhibitor (3r) suppressed the proliferation and invasion of thyroid cancer cells in vitro and vivo. Moreover, FAM20C is driven by KLF12 through its enhancer. Collectively, our study uncovers the potential correlations between the aberrant activation of cancer-specific enhancers and PTC tumorigenesis and identifies FAM20C as a novel target for PTC.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
The ChIP-seq and RNA-seq data generated in this study have been deposited to the NCBI Gene Expression Omnibus (GEO) with the accession codes GSE224355, GSE224356 and Sequence Read Archive (SRA) under accession number PRJNA1070988. The corresponding author can provide all other data to support this study upon reasonable request. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Rossi ED, Pantanowitz L, Hornick JL. A worldwide journey of thyroid cancer incidence centred on tumour histology. Lancet Diabetes Endocrinol. 2021;9:193–4.
Ruan X, Shi X, Dong Q, Yu Y, Hou X, Song X, et al. Antitumor effects of anlotinib in thyroid cancer. Endocr Relat Cancer. 2019;26:153–64.
Du L, Zhao Z, Zheng R, Li H, Zhang S, Li R, et al. Epidemiology of thyroid cancer: incidence and mortality in China, 2015. Front Oncol. 2020;10:1702.
Schneider DF, Chen H. New developments in the diagnosis and treatment of thyroid cancer. CA Cancer J Clin. 2013;63:374–94.
Ruan XH, Tian MR, Kang N, Ma WK, Zeng Y, Zhuang GJ, et al. Genome-wide identification of m6A-associated functional SNPs as potential functional variants for thyroid cancer. Am J Cancer Res. 2021;11:5402–14.
Hou XK, Shi XL, Zhang W, Li DP, Hu LF, Yang JH, et al. LDHA induces EMT gene transcription and regulates autophagy to promote the metastasis and tumorigenesis of papillary thyroid carcinoma. Cell Death Dis. 2021;12:347.
Toraih EA, Hussein MH, Zerfaoui M, Attia AS, Ellythy AM, Mostafa A, et al. Site-specific metastasis and survival in papillary thyroid cancer: the importance of brain and multi-organ disease. Cancers. 2021;13:1625.
Brown AP, Chen J, Hitchcock YJ, Szabo A, Shrieve DC, Tward JD. The risk of second primary malignancies up to three decades after the treatment of differentiated thyroid cancer. J Clin Endocrinol Metab. 2008;93:504–15.
Xing MZ, Haugen BR, Schlumberger M. Progress in molecular-based management of differentiated thyroid cancer. Lancet. 2013;381:1058–69.
Fagin JA, Matsuo K, Karmakar A, Chen DL, Tang SH, Koeffler HP. High prevalence of mutations of the p53 gene in poorly differentiated human thyroid carcinomas. J Clin Invest. 1993;91:179–84.
Donghi R, Longoni A, Pilotti S, Michieli P, Della Porta G, Pierotti MA. Gene p53 mutations are restricted to poorly differentiated and undifferentiated carcinomas of the thyroid gland. J Clin Invest. 1993;91:1753–60.
Murugan AK, Xing M. Anaplastic thyroid cancers harbor novel oncogenic mutations of the ALK gene. Cancer Res. 2011;71:4403–11.
Li S, Hu G, Chen Y, Sang Y, Tang Q, Liu R. TERT upstream promoter methylation regulates TERT expression and acts as a therapeutic target in TERT promoter mutation-negative thyroid cancer. Cancer Cell Int. 2024;24:271.
Liu R, Zhu G, Tan J, Shen X, Xing M. Genetic trio of BRAF and TERT alterations and rs2853669TT in papillary thyroid cancer aggressiveness. J Natl Cancer Inst. 2024;116:694–701.
Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357:eaal2380.
Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336:736–9.
Qamra A, Xing M, Padmanabhan N, Kwok JJT, Zhang S, Xu C, et al. Epigenomic promoter alterations amplify gene isoform and immunogenic diversity in gastric adenocarcinoma. Cancer Discov. 2017;7:630–51.
Yao X, Tan J, Lim KJ, Koh J, Ooi WF, Li Z, et al. VHL deficiency drives enhancer activation of oncogenes in clear cell renal cell carcinoma. Cancer Discov. 2017;7:1284–305.
Lomberk G, Dusetti N, Iovanna J, Urrutia R. Emerging epigenomic landscapes of pancreatic cancer in the era of precision medicine. Nat Commun. 2019;10:3875.
Tang F, Yang Z, Tan Y, Li Y. Super-enhancer function and its application in cancer targeted therapy. NPJ Precis Oncol. 2020;4:2.
Huang H, Hu J, Maryam A, Huang Q, Zhang Y, Ramakrishnan S, et al. Defining super-enhancer landscape in triple-negative breast cancer by multiomic profiling. Nat Commun. 2021;12:2242.
Ye B, Fan D, Xiong W, Li M, Yuan J, Jiang Q, et al. Oncogenic enhancers drive esophageal squamous cell carcinogenesis and metastasis. Nat Commun. 2021;12:4457.
Kelly MR, Wisniewska K, Regner MJ, Lewis MW, Perreault AA, Davis ES, et al. A multi-omic dissection of super-enhancer driven oncogenic gene expression programs in ovarian cancer. Nat Commun. 2022;13:4247.
Zhang L, Xiong D, Liu Q, Luo Y, Tian Y, Xiao X, et al. Genome-wide histone H3K27 acetylation profiling identified genes correlated with prognosis in papillary thyroid carcinoma. Front Cell Dev Biol. 2021;9:682561.
Cao XY, Dang L, Zheng XQ, Lu Y, Lu YM, Ji RJ, et al. Targeting super-enhancer-driven oncogenic transcription by CDK7 inhibition in anaplastic thyroid carcinoma. Thyroid. 2019;29:809–23.
Huang J, Liang X, Xuan Y, Geng C, Li Y, Lu H, et al. A reference human genome dataset of the BGISEQ-500 sequencer. Gigascience. 2017;6:1–9.
Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2018;7:1–6.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–4.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137.
Kramer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–30.
Yokoyama Y, Zhu H, Lee JH, Kossenkov AV, Wu SY, Wickramasinghe JM, et al. BET inhibitors suppress ALDH activity by targeting ALDH1A1 super-enhancer in ovarian cancer. Cancer Res. 2016;76:6320–30.
Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34.
Yamane K, Ihn H, Asano Y, Jinnin M, Tamaki K. Antagonistic effects of TNF-alpha on TGF-beta signaling through down-regulation of TGF-beta receptor type II in human dermal fibroblasts. J Immunol. 2003;171:3855–62.
Szondy Z, Pallai A. Transmembrane TNF-alpha reverse signaling leading to TGF-beta production is selectively activated by TNF targeting molecules: therapeutic implications. Pharmacol Res. 2017;115:124–32.
Liu ZW, Zhang YM, Zhang LY, Zhou T, Li YY, Zhou GC, et al. Duality of interactions between TGF-beta and TNF-alpha during tumor formation. Front Immunol. 2021;12:810286.
Zhao RY, Fu LL, Yuan ZX, Liu Y, Zhang K, Chen YM, et al. Discovery of a novel small-molecule inhibitor of Fam20C that induces apoptosis and inhibits migration in triple negative breast cancer. Eur J Med Chem. 2021;210:113088.
Qin ZY, Wang PQ, Li XY, Zhang SY, Tian M, Dai Y, et al. Systematic network-based discovery of a Fam20C inhibitor (FL-1607) with apoptosis modulation in triple-negative breast cancer. Mol Biosyst. 2016;12:2108–18.
Tagliabracci VS, Engel JL, Wen JZ, Wiley SE, Worby CA, Kinch LN, et al. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science. 2012;336:1150–3.
Ishikawa HO, Xu AG, Ogura E, Manning G, Irvine KD, The raine syndrome protein FAM20C is a golgi kinase that phosphorylates bio-mineralization proteins. PLoS ONE. 2012;7:e42988.
Tagliabracci VS, Wiley SE, Guo X, Kinch LN, Durrant E, Wen JZ, et al. A single kinase generates the majority of the secreted phosphoproteome. Cell. 2015;161:1619–32.
Liu XP, Zhan YB, Xu WX, Liu XY, Geng YW, Liu LX, et al. Prognostic and immunological role of Fam20C in pan-cancer. Biosci Rep. 2021;41:Bsr20201920.
Wu Y, Wang H, Liu C. From biomineralization to tumorogenesis-the expanding insight of the physiological and pathological roles of Fam20C. Biosci Rep. 2021;41:BSR20210040.
Lin B, Jiang X, Bhandari A, Chen Q, Pan Y. FAM20C promotes papillary thyroid cancer proliferation and metastasis via epithelial-mesenchymal transition. Mol Carcinog. 2025;64:152–61.
Nakamura Y, Migita T, Hosoda F, Okada N, Gotoh M, Arai Y, et al. Krüppel-like factor 12 plays a significant role in poorly differentiated gastric cancer progression. Int J Cancer. 2009;125:1859–67.
Pan X, Zhang W, Wang L, Guo H, Zheng M, Wu H, et al. KLF12 transcriptionally regulates PD-L1 expression in non-small cell lung cancer. Mol Oncol. 2023;17:2659–74.
Plenchette S, Romagny S, Laurens V, Bettaieb A. S-Nitrosylation in TNF superfamily signaling pathway: implication in cancer. Redox Biol. 2015;6:507–15.
Wang X, Lin Y. Tumor necrosis factor and cancer, buddies or foes? Acta Pharm Sin. 2008;29:1275–88.
Cruceriu D, Baldasici O, Balacescu O, Berindan-Neagoe I. The dual role of tumor necrosis factor-alpha (TNF-alpha) in breast cancer: molecular insights and therapeutic approaches. Cell Oncol. 2020;43:1–18.
Bauerle KT, Schweppe RE, Haugen BR. Inhibition of nuclear factor-kappa B differentially affects thyroid cancer cell growth, apoptosis, and invasion. Mol Cancer. 2010;9:117.
Mitsiades N, Poulaki V, Tseleni-Balafouta S, Koutras DA, Stamenkovic I. Thyroid carcinoma cells are resistant to FAS-mediated apoptosis but sensitive to tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 2000;60:4122–9.
Pisarev MA, Thomasz L, Juvenal GJ. Role of transforming growth factor beta in the regulation of thyroid function and growth. Thyroid. 2009;19:881–92.
Zhi J, Zhang P, Zhang W, Ruan X, Tian M, Guo S, et al. Inhibition of BRAF sensitizes thyroid carcinoma to immunotherapy by enhancing tsMHCII-mediated immune recognition. J Clin Endocrinol Metab. 2021;106:91–107.
Dash S, Sahu AK, Srivastava A, Chowdhury R, Mukherjee S. Exploring the extensive crosstalk between the antagonistic cytokines- TGF-beta and TNF-alpha in regulating cancer pathogenesis. Cytokine. 2021;138:155348.
Kalliolias GD, Ivashkiv LB. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat Rev Rheumatol. 2016;12:49–62.
Nikiforov YE. Thyroid carcinoma: molecular pathways and therapeutic targets. Mod Pathol. 2008;21:S37–S43.
Naoum GE, Morkos M, Kim B, Arafat W. Novel targeted therapies and immunotherapy for advanced thyroid cancers. Mol Cancer. 2018;17:51.
Hu LF, Zhang J, Tian MR, Kang N, Xu GW, Zhi JT, et al. Pharmacological inhibition of Ref-1 enhances the therapeutic sensitivity of papillary thyroid carcinoma to vemurafenib. Cell Death Dis. 2022;13:124.
Acknowledgements
We thank Prof. Yupeng Chen (Tianjin Medical University) and Prof. Nan Qin (Tianjin Medical University) for generously providing technical support and helpful discussion and suggestions.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82172821, 82103386, 82272721, 82103234), Tianjin Municipal Science and Technology Project (19JCYBJC27400, 21JCZDJC00360), and Beijing-Tianjin-Hebei Basic Research Cooperation Project (20JCZXJC00120), The Science & Technology Development Fund of Tianjin Education Commission for Higher Education (2021ZD033), Tianjin Medical Key Discipline (Specialty) Construction Project (TJYXZDXK-058B), Tianjin Health Research Project (TJWJ2022XK024).
Author information
Authors and Affiliations
Contributions
Xiangqian Zheng: writing—review & editing, writing—original draft, validation, software, resources, methodology, investigation, funding acquisition. Xianhui Ruan: writing—review & editing, validation, software, resources, methodology, investigation, data curation, conceptualisation, funding acquisition. Wei Zhang: writing—review & editing, resources, methodology, investigation, conceptualisation. Xiukun Hou: writing—original draft, validation, software, investigation, formal analysis, data curation. Guiming Fu: resources. Weike Ma: supervision, software, resources, investigation. Jianfeng Huang: writing—original draft, supervision, resources, methodology, investigation. Yuyang Qian: software, project administration, investigation, data curation. Mengran Tian: software, methodology, formal analysis, data curation. Nan Qin: validation, software, investigation. Yupeng Chen: writing—review & editing. Ming Gao: writing—review & editing, supervision, project administration, funding acquisition, conceptualisation. Dapeng Li: writing—review & editing, validation, supervision, project administration, funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Tianjin Medical University Cancer Institute and Hospital (EK2021149), and was performed according to the Declaration of Helsinki.
Consent for publication
All co-authors have read and approved the final version of the manuscript and its submission to this journal.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ruan, X., Zhang, W., Hou, X. et al. Enhancer hijacking drives FAM20C expression to promote papillary thyroid cancer progression. Cancer Gene Ther 32, 854–869 (2025). https://doi.org/10.1038/s41417-025-00930-8
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41417-025-00930-8