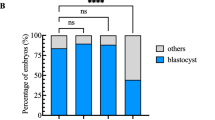
Abstract
Zygotic chromatin undergoes extensive reprogramming immediately after fertilization. It is generally accepted that maternal factors control this process. However, little is known about the underlying mechanisms. Here we report that maternal RAD9A, a key protein in DNA damage response pathway, is involved in post-zygotic embryo development, via a mouse model with conditional depletion of Rad9a alleles in oocytes of primordial follicles. Post-zygotic losses originate from delayed zygotic chromatin decondensation after depletion of maternal RAD9A. Pronucleus formation and DNA replication of most mutant zygotes are therefore deferred, which subsequently trigger the G2/M checkpoint and arrest development of most mutant zygotes. Delayed zygotic chromatin decondensation could also lead to increased reabsorption of post-implantation mutant embryos. In addition, our data indicate that delayed zygotic chromatin decondensation may be attributed to deferred epigenetic modification of histone in paternal chromatin after fertilization, as fertilization and resumption of secondary meiosis in mutant oocytes were both normal. More interestingly, most mutant oocytes could not support development beyond one-cell stage after parthenogenetic activation. Therefore, RAD9A may also play an important role in maternal chromatin reprogramming. In summary, our data reveal an important role of RAD9A in zygotic chromatin reprogramming and female fertility.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Clift D, Schuh M. Restarting life: fertilization and the transition from meiosis to mitosis. Nat Rev Mol Cell Biol. 2013;14:549–62.
Cabot B, Cabot RA. Chromatin remodeling in mammalian embryos. Reproduction. 2018;155:R147–R158.
Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–10.
Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–12.
Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell. 2014;30:268–79.
Du Z, Zheng H, Huang B, Ma R, Wu J, Zhang X, et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 2017;547:232–5.
Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–7.
Niida H, Nakanishi M. DNA damage checkpoints in mammals. Mutagenesis. 2006;21:3–9.
Jekimovs C, Bolderson E, Suraweera A, Adams M, O’Byrne KJ, Richard DJ. Chemotherapeutic compounds targeting the DNA double-strand break repair pathways: the good, the bad, and the promising. Front Oncol. 2014;4:86.
Bashkirov VI, Bashkirova EV, Haghnazari E, Heyer WD. Direct kinase-to-kinase signaling mediated by the FHA phosphoprotein recognition domain of the Dun1 DNA damage checkpoint kinase. Mol Cell Biol. 2003;23:1441–52.
Lieberman HB. Rad9, an evolutionarily conserved gene with multiple functions for preserving genomic integrity. J Cell Biochem. 2006;97:690–7.
Griffith JD, Lindsey-Boltz LA, Sancar A. Structures of the human Rad17-replication factor C and checkpoint Rad 9-1-1 complexes visualized by glycerol spray/low voltage microscopy. J Biol Chem. 2002;277:15233–6.
Dang T, Bao S, Wang XF. Human Rad9 is required for the activation of S-phase checkpoint and the maintenance of chromosomal stability. Genes Cells. 2005;10:287–95.
Pandita RK, Sharma GG, Laszlo A, Hopkins KM, Davey S, Chakhparonian M, et al. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol Cell Biol. 2006;26:1850–64.
Vasileva A, Hopkins KM, Wang X, Weisbach MM, Friedman RA, Wolgemuth DJ, et al. The DNA damage checkpoint protein RAD9A is essential for male meiosis in the mouse. J Cell Sci. 2013;126(Pt 17):3927–38.
Yin Y, Zhu A, Jin YJ, Liu YX, Zhang X, Hopkins KM, et al. Human RAD9 checkpoint control/proapoptotic protein can activate transcription of p21. Proc Natl Acad Sci USA. 2004;101:8864–9.
Komatsu K, Miyashita T, Hang H, Hopkins KM, Zheng W, Cuddeback S, et al. Human homologue of S. pombe Rad9 interacts with BCL-2/BCL-xL and promotes apoptosis. Nat Cell Biol. 2000;2:1–6.
Lee MW, Hirai I, Wang HG. Caspase-3-mediated cleavage of Rad9 during apoptosis. Oncogene. 2003;22:6340–6.
Huang L, Wang ZB, Qi ST, Ma XS, Liang QX, Lei G, et al. Rad9a is required for spermatogonia differentiation in mice. Oncotarget. 2016;7:86350–8.
Zhu A, Zhang CX, Lieberman HB. Rad9 has a functional role in human prostate carcinogenesis. Cancer Res. 2008;68:1267–74.
Broustas CG, Lieberman HB. Contributions of Rad9 to tumorigenesis. J Cell Biochem. 2012;113:742–51.
Hu Z, Liu Y, Zhang C, Zhao Y, He W, Han L, et al. Targeted deletion of Rad9 in mouse skin keratinocytes enhances genotoxin-induced tumor development. Cancer Res. 2008;68:5552–61.
Cheng CK, Chow LW, Loo WT, Chan TK, Chan V. The cell cycle checkpoint gene Rad9 is a novel oncogene activated by 11q13 amplification and DNA methylation in breast cancer. Cancer Res. 2005;65:8646–54.
Hopkins KM, Auerbach W, Wang XY, Hande MP, Hang H, Wolgemuth DJ, et al. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol Cell Biol. 2004;24:7235–48.
Kleiman NJ, David J, Elliston CD, Hopkins KM, Smilenov LB, Brenner DJ, et al. Mrad9 and atm haploinsufficiency enhance spontaneous and X-ray-induced cataractogenesis in mice. Radiat Res. 2007;168:567–73.
Lan ZJ, Xu X, Cooney AJ. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol Reprod. 2004;71:1469–74.
Hu MW, Wang ZB, Schatten H, Sun QY. New understandings on folliculogenesis/oogenesis regulation in mouse as revealed by conditional knockout. J Genet Genom. 2012;39:61–68.
Smilenov LB, Lieberman HB, Mitchell SA, Baker RA, Hopkins KM, Hall EJ. Combined haploinsufficiency for ATM and RAD9 as a factor in cell transformation, apoptosis, and DNA lesion repair dynamics. Cancer Res 2005;65:933–8.
Yan L, Donze JR, Liu L. Inactivated MGMT by O6-benzylguanine is associated with prolonged G2/M arrest in cancer cells treated with BCNU. Oncogene. 2005;24:2175–83.
Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–90.
Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280:225–36.
Ma XS, Chao SB, Huang XJ, Lin F, Qin L, Wang XG, et al. The dynamics and regulatory mechanism of pronuclear H3k9me2 asymmetry in mouse zygotes. Sci Rep. 2015;5:17924.
Crichton JH, Playfoot CJ, Adams IR. The role of chromatin modifications in progression through mouse meiotic prophase. J Genet Genom. 2014;41:97–106.
Greer DA, Besley BD, Kennedy KB, Davey S. hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II beta binding protein 1 focus formation. Cancer Res. 2003;63:4829–35.
Ma XS, Lin F, Wang ZW, Hu MW, Huang L, Meng TG, et al. Geminin deletion in mouse oocytes results in impaired embryo development and reduced fertility. Mol Biol Cell. 2016;27:768–75.
Chao SB, Guo L, Ou XH, Luo SM, Wang ZB, Schatten H, et al. Heated spermatozoa: effects on embryonic development and epigenetics. Hum Reprod. 2012;27:1016–24.
Guo L, Chao SB, Xiao L, Wang ZB, Meng TG, Li YY, et al. Sperm-carried RNAs play critical roles in mouse embryonic development. Oncotarget. 2017;8:67394–405.
Jiang ZZ, Hu MW, Wang ZB, Huang L, Lin F, Qi ST, et al. Survivin is essential for fertile egg production and female fertility in mice. Cell Death Dis. 2014;5:e1154.
Qi ST, Wang ZB, Ouyang YC, Zhang QH, Hu MW, Huang X, et al. Overexpression of SETbeta, a protein localizing to centromeres, causes precocious separation of chromatids during the first meiosis of mouse oocytes. J Cell Sci. 2013;126:1595–603.
Hu MW, Wang ZB, Jiang ZZ, Qi ST, Huang L, Liang QX, et al. Scaffold subunit Aalpha of PP2A is essential for female meiosis and fertility in mice. Biol Reprod. 2014;91:19.
Wang ZB, Jiang ZZ, Zhang QH, Hu MW, Huang L, Ou XH, et al. Specific deletion of Cdc42 does not affect meiotic spindle organization/migration and homologous chromosome segregation but disrupts polarity establishment and cytokinesis in mouse oocytes. Mol Biol Cell. 2013;24:3832–41.
Zhang D, Ma W, Li YH, Hou Y, Li SW, Meng XQ, et al. Intra-oocyte localization of MAD2 and its relationship with kinetochores, microtubules, and chromosomes in rat oocytes during meiosis. Biol Reprod. 2004;71:740–8.
Acknowledgements
We are grateful to Shi-Wen Li, Li-Juan Wang, and Hua Qin for their technical assistance. We also thank Dr. Heng-Yu Fan for providing antibodies, Dr. Enkui Duan, Dr. Ying Zhang, Dr. Zhi-Ming Han, and Dr. Jing-Pian Peng for their useful suggestions. This study was supported by the National Key R& D Program of China (2016YFA0100400, 2016YFC1000600) and the National Natural Science Foundation of China (No. 31272260, 31371451, 31530049).
Author information
Authors and Affiliations
Contributions
L.H. and Q.-Y.S. conceived and designed the experiments. L.H., T.-G.M., X.-S.M., Z.-B.W., S.-T.Q., Q.C., Q.-H.Z., Q.-X.L., Z.-W.W., M.-W.H., L.G., and Y.-C.O. performed the experiments. L.H. analyzed the data. Y.H. contributed reagents, materials and analysis tools. Y.Z. provided the Rad9aF/F mouse strain. L.H. and Q.-Y.S. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by M Piacetini.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Huang, L., Meng, TG., Ma, XS. et al. Rad9a is involved in chromatin decondensation and post-zygotic embryo development in mice. Cell Death Differ 26, 969–980 (2019). https://doi.org/10.1038/s41418-018-0181-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-018-0181-9