Abstract
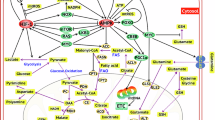
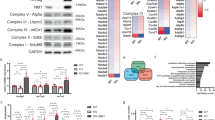
Cancer cells are known to upregulate aerobic glycolysis to promote growth, proliferation, and survival. However, the role of mitochondrial respiration in tumorigenesis remains elusive. Here we report that inhibition of mitochondrial function by silencing TFAM, a key transcription factor essential for mitochondrial DNA (mtDNA) replication and the transcription of mtDNA-encoded genes, markedly reduced tumor-initiating potential of colon cancer cells. Knockdown of TFAM significantly decreased mitochondrial respiration in colon cancer cells; however, the cellular levels of ATP remained largely unchanged as a result of increased glycolysis. This metabolic alteration rendered cancer cells highly susceptible to glucose deprivation. Interestingly, upregulation of glycolysis was independent of hypoxia-inducible factor-1 (HIF1) as TFAM knockdown cells fail to stabilize HIF1α under hypoxic conditions. Moreover, knockdown of TFAM results in decreased expression of genes-associated cancer stem cells downstream of Wnt/β-catenin signaling. Metabolic analysis reveals that the level of α-ketoglutarate (α-KG) was significantly upregulated in TFAM knockout cells. Silencing of prolyl hydroxylase domain-containing protein 2 (PHD2), a α-KG-dependent dioxyenase, rescued the expression of target genes of both HIF1α and Wnt/β-catenin. Furthermore, intestinal-specific knockout of TFAM prevents tumor formation in Apc-mutant mouse models of colon cancer. Taken together, our findings identify a novel role of mitochondria-mediated retrograde signaling in regulating Wnt signaling and tumor initiation in colon cancer.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33.
Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol. 2015;11:9–15.
Chandel NS. Mitochondria as signaling organelles. BMC Biol. 2014;12:34.
DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200.
Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet. 2012;13:878–90.
Kang D, Kim SH, Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions. Mitochondrion. 2007;7:39–44.
Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203.
Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–44.
Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–6.
Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci USA. 2010;107:8788–93.
Janiszewska M, Suva ML, Riggi N, Houtkooper RH, Auwerx J, Clement-Schatlo V, et al. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012;26:1926–44.
Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, et al. MYC/PGC-1alpha balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015;22:590–605.
Vlashi E, Lagadec C, Vergnes L, Matsutani T, Masui K, Poulou M, et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proc Natl Acad Sci USA. 2011;108:16062–7.
Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–32.
Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–13.
Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80.
Vermeulen L, Morrissey E, van der Heijden M, Nicholson AM, Sottoriva A, Buczacki S, et al. Defining stem cell dynamics in models of intestinal tumor initiation. Science. 2013;342:995–8.
Zeuner A, Todaro M, Stassi G, De Maria R. Colorectal cancer stem cells: from the crypt to the clinic. Cell Stem Cell. 2014;15:692–705.
Wen YA, Xiong X, Harris JW, Zaytseva YY, Mitov MI, Napier DL, et al. Adipocytes activate mitochondrial fatty acid oxidation and autophagy to promote tumor growth in colon cancer. Cell death Dis. 2017;8:e2593.
Hamanaka RB, Glasauer A, Hoover P, Yang S, Blatt H, Mullen AR, et al. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Sci Signal. 2013;6:ra8.
Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5:343–54.
Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308.
Cairns RA, Harris I, McCracken S, Mak TW. Cancer cell metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:299–311.
Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–34.
Mancini R, Noto A, Pisanu ME, De Vitis C, Maugeri-Sacca M, Ciliberto G. Metabolic features of cancer stem cells: the emerging role of lipid metabolism. Oncogene. 2018;37:2367–2378.
Song IS, Jeong YJ, Jeong SH, Heo HJ, Kim HK, Bae KB, et al. FOXM1-induced PRX3 regulates stemness and survival of colon cancer cells via maintenance of mitochondrial function. Gastroenterology. 2015;149:1006–16 e9.
Srivillibhuthur M, Warder BN, Toke NH, Shah PP, Feng Q, Gao N, et al. TFAM is required for maturation of the fetal and adult intestinal epithelium. Dev Biol. 2018;439:92–101.
Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481:385–8.
Woo DK, Green PD, Santos JH, D’Souza AD, Walther Z, Martin WD, et al. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC(Min/ +) mice. Am J Pathol. 2012;180:24–31.
Hamanaka RB, Weinberg SE, Reczek CR, Chandel NS. The mitochondrial respiratory chain is required for organismal adaptation to hypoxia. Cell Rep. 2016;15:451–9.
Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51:794–8.
Panieri E, Santoro MM. ROS homeostasis and metabolism: a dangerous liason in cancer cells. Cell death Dis. 2016;7:e2253.
Watanabe A, Arai M, Koitabashi N, Niwano K, Ohyama Y, Yamada Y, et al. Mitochondrial transcription factors TFAM and TFB2M regulate Serca2 gene transcription. Cardiovasc Res. 2011;90:57–67.
Pastukh V, Shokolenko I, Wang B, Wilson G, Alexeyev M. Human mitochondrial transcription factor A possesses multiple subcellular targeting signals. FEBS J. 2007;274:6488–99.
Wang YE, Marinov GK, Wold BJ, Chan DC. Genome-wide analysis reveals coating of the mitochondrial genome by TFAM. PLoS One. 2013;8:e74513.
Li X, Stevens PD, Liu J, Yang H, Wang W, Wang C, et al. PHLPP is a negative regulator of RAF1, which reduces colorectal cancer cell motility and prevents tumor progression in mice. Gastroenterology. 2014;146:1301–12. e1-10
Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene. 2009;28:994–1004.
Cheung AF, Carter AM, Kostova KK, Woodruff JF, Crowley D, Bronson RT, et al. Complete deletion of Apc results in severe polyposis in mice. Oncogene. 2010;29:1857–64.
Wen YA, Xiong X, Zaytseva YY, Napier DL, Vallee E, Li AT, et al. Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell death Dis. 2018;9:265.
Xiong X, Wen YA, Mitov MI, M CO, Miyamoto S, Gao T. PHLPP regulates hexokinase 2-dependent glucose metabolism in colon cancer cells. Cell Death Discov. 2017;3:16103.
Sun RC, Fan TW, Deng P, Higashi RM, Lane AN, Le AT, et al. Noninvasive liquid diet delivery of stable isotopes into mouse models for deep metabolic network tracing. Nat Commun. 2017;8:1646.
Fan TW, Lane AN. Assignment strategies for NMR resonances in metabolomics research (eds. Lutz NW, Sweedler JV and Wevers RA). In: Methodologies for metabolomics: experimental strategies and techniques. New York: Cambridge University Press; 2013. p. 525–84.
Fan TW, Lane AN. Structure-based profiling of metabolites and isotopomers by NMR. Prog Nucl Magn Reson Spectrosc. 2008;52:69–117.
Fan TW, Lane AN. NMR-based stable isotope resolved metabolomics in systems biochemistry. J Biomol NMR. 2011;49:267–80.
Lane AN, Fan TW, Higashi RM. Isotopomer-based metabolomic analysis by NMR and mass spectrometry. Methods Cell Biol. 2008;84:541–88.
Fan TW, Warmoes MO, Sun Q, Song H, Turchan-Cholewo J, Martin JT, et al. Distinctly perturbed metabolic networks underlie differential tumor tissue damages induced by immune modulator beta-glucan in a two-case ex vivo non-small-cell lung cancer study. Cold Spring Harb Mol Case Stud. 2016;2:a000893.
Moseley HN. Correcting for the effects of natural abundance in stable isotope resolved metabolomics experiments involving ultra-high resolution mass spectrometry. BMC Bioinformatics. 2010;11:139.
Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Acknowledgements
This work was supported by R01CA133429 (TG), 1U24DK097215 (T.W.-M.F.) and 5R35CA197532 (NSC). The studies were conducted with support provided by the Redox Metabolism, Biospecimen Procurement and Translational Pathology, Flow Cytometry and Cell Sorting, and Biostatistics and Bioinformatics Shared Resource Facilities of the University of Kentucky Markey Cancer Center (P30CA177558) as well as the Center for Environmental and Systems Biochemistry (CESB) at the University of Kentucky.
Author information
Authors and Affiliations
Contributions
Y-AW, TAB, and TG designed all aspects of the study; Y-AW, XX, TS, ATL, CW, HLW, and LT performed experiments and analysis; EB, TWMF, and NSC provided input into the project; Y-AW and TG wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by S. Kaufmann
Supplementary information
Rights and permissions
About this article
Cite this article
Wen, YA., Xiong, X., Scott, T. et al. The mitochondrial retrograde signaling regulates Wnt signaling to promote tumorigenesis in colon cancer. Cell Death Differ 26, 1955–1969 (2019). https://doi.org/10.1038/s41418-018-0265-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-018-0265-6
This article is cited by
-
Total cardiolipin levels in gastric and colon cancer: evaluating the prognostic potential
Lipids in Health and Disease (2025)
-
Regulatory effect of Wnt signaling on mitochondria in cancer: from mechanism to therapy
Apoptosis (2025)
-
Integrated control of cancer stemness by σ1 receptor in advanced prostate cancer
Oncogene (2025)
-
Characterization of the stem cell landscape and identification of a stemness-associated prognostic signature in bladder cancer
Cancer Cell International (2024)
-
Mitochondrial function and gastrointestinal diseases
Nature Reviews Gastroenterology & Hepatology (2024)