Abstract
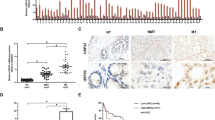
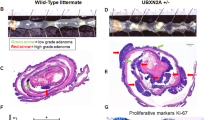
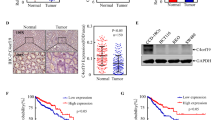
USP22, the deubiquitinating subunit of the SAGA transcriptional cofactor complex, is a member of an 11-gene “death-from-cancer” signature. USP22 has been considered an attractive therapeutic target since high levels of its expression were associated with distant metastasis, poor survival, and high recurrence rates in a wide variety of solid tumors, including colorectal cancer (CRC). We sought to investigate the role of Usp22 during tumorigenesis in vivo using a mouse model for intestinal carcinogenesis with a tissue-specific Usp22 ablation. In addition, we assessed the effects of USP22 depletion in human CRC cells on tumorigenic potential and identified underlying molecular mechanisms. For the first time, we report that USP22 has an unexpected tumor-suppressive function in vivo. Intriguingly, intestine-specific Usp22 deletion exacerbated the tumor phenotype caused by Apc mutation, resulting in significantly decreased survival and higher intestinal tumor incidence. Accordingly, human CRC cells showed increased tumorigenic properties upon USP22 reduction in vitro and in vivo and induced gene expression signatures associated with an unfavorable outcome in CRC patients. Notably, USP22 loss resulted in increased mTOR activity with the tumorigenic properties elicited by the loss of USP22 being reversible by mTOR inhibitor treatment in vitro and in vivo. Here, we demonstrate that USP22 can exert tumor-suppressive functions in CRC where its loss increases CRC burden by modulating mTOR activity. Importantly, our data uncover a tumor- and context-specific role of USP22, suggesting that USP22 expression could serve as a marker for therapeutic stratification of cancer patients.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bosetti C, Levi F, Rosato V, Bertuccio P, Lucchini F, Negri E, et al. Recent trends in colorectal cancer mortality in Europe. Int J Cancer. 2011;129:180–91. https://doi.org/10.1002/ijc.25653
Vleugels JLA, van Lanschot MCJ, Dekker E. Colorectal cancer screening by colonoscopy: putting it into perspective. Dig Endosc. 2016;28:250–9. https://doi.org/10.1111/den.12533
Glinsky GV. Death-from-cancer signatures and stem cell contribution to metastatic cancer. Cell Cycle. 2005;4:1171–5. https://doi.org/10.4161/cc.4.9.2001
Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Investig. 2005;115:1503–21. https://doi.org/10.1172/JCI23412
Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea–a paradigm shift. Cancer Res. 2006;66:1883–90. https://doi.org/10.1158/0008-5472.CAN-05-3153. discussion 1895-6
Zhang X-Y, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, et al. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–11. https://doi.org/10.1016/j.molcel.2007.12.015
Zhang X-Y, Pfeiffer HK, Thorne AW, McMahon SB. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle. 2008;7:1522–4. https://doi.org/10.4161/cc.7.11.5962
Atanassov BS, Mohan RD, Lan X, Kuang X, Lu Y, Lin K, et al. ATXN7L3 and ENY2 coordinate activity of multiple H2B deubiquitinases important for cellular proliferation and tumor growth. Mol Cell. 2016;62:558–71. https://doi.org/10.1016/j.molcel.2016.03.030
Li L, Osdal T, Ho Y, Chun S, McDonald T, Agarwal P, et al. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell Stem Cell. 2014;15:431–46. https://doi.org/10.1016/j.stem.2014.08.001
Lin Z, Yang H, Kong Q, Li J, Lee S-M, Gao B, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell. 2012;46:484–94. https://doi.org/10.1016/j.molcel.2012.03.024
Ding F, Bao C, Tian Y, Xiao H, Wang M, Xie X, et al. USP22 promotes NSCLC tumorigenesis via MDMX up-regulation and subsequent p53 inhibition. Int J Mol Sci. 2014;16:307–20. https://doi.org/10.3390/ijms16010307
Gao Y, Lin F, Xu P, Nie J, Chen Z, Su J, et al. USP22 is a positive regulator of NFATc2 on promoting IL2 expression. FEBS Lett. 2014;588:878–83. https://doi.org/10.1016/j.febslet.2014.02.016
Melling N, Grimm N, Simon R, Stahl P, Bokemeyer C, Terracciano L, et al. Loss of H2Bub1 expression is linked to poor prognosis in nodal negative colorectal cancers. Pathol Oncol Res. 2016;22:95–102. https://doi.org/10.1007/s12253-015-9977-9
Liu YL, Yang YM, Xu H, Dong XS. Increased expression of ubiquitin-specific protease 22 can promote cancer progression and predict therapy failure in human colorectal cancer. J Gastroenterol Hepatol. 2010;25:1800–5. https://doi.org/10.1111/j.1440-1746.2010.06352.x
Liu Y, Yang Y, Xu H, Dong X. Implication of USP22 in the regulation of BMI-1, c-Myc, p16INK4a, p14ARF, and cyclin D2 expression in primary colorectal carcinomas. Diagn Mol Pathol. 2010;19:194–200. https://doi.org/10.1097/PDM.0b013e3181e202f2
Liu Y-L, Yang Y-M, Xu H, Dong X-S. Aberrant expression of USP22 is associated with liver metastasis and poor prognosis of colorectal cancer. J Surg Oncol. 2011;103:283–9. https://doi.org/10.1002/jso.21802
Jiang S, Song C, Gu X, Wang M, Miao D, Lv J, et al. Ubiquitin-specific peptidase 22 contributes to colorectal cancer stemness and chemoresistance via Wnt/β-catenin pathway. Cell Physiol Biochem. 2018;46:1412–22. https://doi.org/10.1159/000489156
Melo-Cardenas J, Zhang Y, Zhang DD, Fang D. Ubiquitin-specific peptidase 22 functions and its involvement in disease. Oncotarget. 2016;7:44848–56. https://doi.org/10.18632/oncotarget.8602
Koutelou E, Wang L, Schibler AC, Chao H-P, Kuang X, Lin K, et al. USP22 controls multiple signaling pathways that are essential for vasculature formation in the mouse placenta. Development. 2019. https://doi.org/10.1242/dev.174037.
Jeusset LM-P, McManus KJ. Ubiquitin specific peptidase 22 regulates histone H2B mono-ubiquitination and exhibits both oncogenic and tumor suppressor roles in cancer. Cancers. 2017. https://doi.org/10.3390/cancers9120167
Cremers N, Neeb A, Uhle T, Dimmler A, Rothley M, Allgayer H, et al. CD24 is not required for tumor initiation and growth in murine breast and prostate cancer models. PLoS ONE. 2016;11:e0151468. https://doi.org/10.1371/journal.pone.0151468
van der Bilt ARM, van Terwisscha Scheltinga AGT, Timmer-Bosscha H, Schröder CP, Pot L, Kosterink JGW, et al. Measurement of tumor VEGF-A levels with 89Zr-bevacizumab PET as an early biomarker for the antiangiogenic effect of everolimus treatment in an ovarian cancer xenograft model. Clin Cancer Res. 2012;18:6306–14. https://doi.org/10.1158/1078-0432.CCR-12-0406
Kosinsky RL, Wegwitz F, Hellbach N, Dobbelstein M, Mansouri A, Vogel T, et al. Usp22 deficiency impairs intestinal epithelial lineage specification in vivo. Oncotarget. 2015;6:37906–18. https://doi.org/10.18632/oncotarget.5412
Kosinsky RL, Chua RL, Qui M, Saul D, Mehlich D, Ströbel P, et al. Loss of RNF40 decreases NF-κB activity in colorectal cancer cells and reduces colitis burden in mice. J Crohns Colitis. 2018. https://doi.org/10.1093/ecco-jcc/jjy165.
Prenzel T, Begus-Nahrmann Y, Kramer F, Hennion M, Hsu C, Gorsler T, et al. Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer Res. 2011;71:5739–53. https://doi.org/10.1158/0008-5472.CAN-11-1896
Schneider D, Chua RL, Molitor N, Hamdan FH, Rettenmeier EM, Prokakis E, et al. The E3 ubiquitin ligase RNF40 suppresses apoptosis in colorectal cancer cells. Clin Epigenetics. 2019;11:98 https://doi.org/10.1186/s13148-019-0698-x
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. https://doi.org/10.1073/pnas.0506580102
Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. https://doi.org/10.1038/ng1180
Fan J-D, Lei P-J, Zheng J-Y, Wang X, Li S, Liu H, et al. The selective activation of p53 target genes regulated by SMYD2 in BIX-01294 induced autophagy-related cell death. PLoS ONE. 2015;10:e0116782. https://doi.org/10.1371/journal.pone.0116782
Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017. https://doi.org/10.1126/science.aan2507
Nordgren S, McPheeters G, Svaninger G, Oresland T, Hultén L. Small bowel length in inflammatory bowel disease. Int J Colorectal Dis. 1997;12:230–4.
Wang X-W, Zhang Y-J. Targeting mTOR network in colorectal cancer therapy. World J Gastroenterol. 2014;20:4178–88. https://doi.org/10.3748/wjg.v20.i15.4178
Melo-Cardenas J, Xu Y, Wei J, Tan C, Kong S, Gao B, et al. USP22 deficiency leads to myeloid leukemia upon oncogenic Kras activation through a PU.1-dependent mechanism. Blood. 2018;132:423–34. https://doi.org/10.1182/blood-2017-10-811760
Acknowledgements
The authors are grateful to N. Molitor and S. Beuermann for their technical assistance and the staff of the animal facility at the European Neuroscience Institute Göttingen (ENI-G). Furthermore, we thank S. Becker for performing FACS at the Cell Sorting Core Facility, Department of Hematology and Medical Oncology, University Medicine Göttingen (UMG), Göttingen, as well as G. Salinas and F. Ludewig for performing next-generation sequencing at the Transcriptome and Genome Analysis Laboratory (TAL) Göttingen. This work was supported by funding from the Roggenbuck Foundation (to RLK) and institutional funding provided to the Department of General, Visceral and Pediatric Surgery by the University Medical Center Göttingen (to SAJ). RLK is supported by the Dorothea Schlözer program (University of Göttingen).
Author information
Authors and Affiliations
Contributions
SAJ and RLK designed the study with input from YB-N. RLK performed all the cell culture experiments with the help of MZ and DS. Next-generation sequencing data were generated and analyzed by RLK and FW provided support with bioinformatic analyses. Mouse experiments were performed by RLK with the help of FW, DS, and MZ. Histological analyses were carried out by RLK and LW. ChIP was performed by RLK and MZ with the help of XW. SAJ and RLK wrote the manuscript. All authors have read and approved the final version of the article, including the authorship list.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Melino
Supplementary information
Rights and permissions
About this article
Cite this article
Kosinsky, R.L., Zerche, M., Saul, D. et al. USP22 exerts tumor-suppressive functions in colorectal cancer by decreasing mTOR activity. Cell Death Differ 27, 1328–1340 (2020). https://doi.org/10.1038/s41418-019-0420-8
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-019-0420-8
This article is cited by
-
Immunoregulatory roles of post-translational modifications in colorectal cancer: mechanisms and therapeutic implications
Cellular and Molecular Life Sciences (2025)
-
USP39 promote post-translational modifiers to stimulate the progress of cancer
Discover Oncology (2025)
-
AMPKα2 promotes tumor immune escape by inducing CD8+ T-cell exhaustion and CD4+ Treg cell formation in liver hepatocellular carcinoma
BMC Cancer (2024)
-
Deubiquitinases in cancer
Nature Reviews Cancer (2023)
-
Phosphorylation of USP27X by GSK3β maintains the stability and oncogenic functions of CBX2
Cell Death & Disease (2023)