Abstract
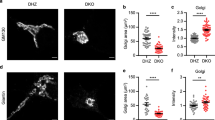
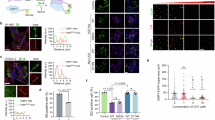
Oleate, the most abundant endogenous and dietary cis-unsaturated fatty acid, has the atypical property to cause the redistribution of microtubule-associated proteins 1A/1B light chain 3B (referred to as LC3) to the trans-Golgi network (TGN), as shown here. A genome-wide screen identified multiple, mostly Golgi transport-related genes specifically involved in the oleate-induced relocation of LC3 to the Golgi apparatus. Follow-up analyses revealed that oleate also caused the retention of secreted proteins in the TGN, as determined in two assays in which the secretion of proteins was synchronized, (i) an assay involving a thermosensitive vesicular stomatitis virus G (VSVG) protein that is retained in the endoplasmic reticulum (ER) until the temperature is lowered, and (ii) an isothermic assay involving the reversible retention of the protein of interest in the ER lumen and that was used both in vitro and in vivo. A pharmacological screen searching for agents that induce LC3 aggregation at the Golgi apparatus led to the identification of “oleate mimetics” that share the capacity to block conventional protein secretion. In conclusion, oleate represents a class of molecules that act on the Golgi apparatus to cause the recruitment of LC3 and to stall protein secretion.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8.
Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, et al. Molecular definitions of autophagy and related processes. The EMBO journal. 2017;36:1811–36.
Mizushima N. The ATG conjugation systems in autophagy. Current Opinion Cell Biol. 2019;63:1–10.
Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26.
Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1–222.
Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–80.
Hansen M, Rubinsztein DC, Walker DW. Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol. 2018;19:579–93.
Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42.
Morishita H, Mizushima N. Diverse cellular roles of autophagy. Annual Rev Cell Dev Biol. 2019;35:453–75.
Sales-Campos H, Souza PR, Peghini BC, da Silva JS, Cardoso CR. An overview of the modulatory effects of oleic acid in health and disease. Mini Rev Med Chem. 2013;13:201–10.
Widmer RJ, Flammer AJ, Lerman LO, Lerman A. The Mediterranean diet, its components, and cardiovascular disease. Am J Med. 2015;128:229–38.
Niso-Santano M, Malik SA, Pietrocola F, Bravo-San Pedro JM, Marino G, Cianfanelli V, et al. Unsaturated fatty acids induce non-canonical autophagy. EMBO J. 2015;34:1025–41.
Sauvat A, Chen G, Muller K, Tong M, Aprahamian F, Durand S, et al. Trans-fats inhibit autophagy induced by saturated fatty acids. EBioMed. 2018;30:261–72.
Sundqvist M, Christenson K, Holdfeldt A, Gabl M, Martensson J, Bjorkman L, et al. Similarities and differences between the responses induced in human phagocytes through activation of the medium chain fatty acid receptor GPR84 and the short chain fatty acid receptor FFA2R. Biochim et biophys acta Mol Cell Res. 2018;1865:695–708.
Bolognini D, Barki N, Butcher AJ, Hudson BD, Sergeev E, Molloy C, et al. Chemogenetics defines receptor-mediated functions of short chain free fatty acids. Nat Chem Biol. 2019;15:489–98.
Luckmann M, Trauelsen M, Frimurer TM, Schwartz TW. Structural basis for GPCR signaling by small polar versus large lipid metabolites-discovery of non-metabolite ligands. Current Opinion Cell Biol. 2020;63:38–48.
de Souza RJ, Mente A, Maroleanu A, Cozma AI, Ha V, Kishibe T, et al. Intake of saturated and trans unsaturated fatty acids and risk of all cause mortality, cardiovascular disease, and type 2 diabetes: systematic review and meta-analysis of observational studies. Bmj. 2015;351:h3978.
Wang DD, Hu FB. Dietary fat and risk of cardiovascular disease: recent controversies and advances. Annual Rev Nutr. 2017;37:423–46.
Corella D, Coltell O, Macian F, Ordovas JM. Advances in understanding the molecular basis of the mediterranean diet effect. Annual Rev Food Sci Techn. 2018;9:227–49.
Enot DP, Niso-Santano M, Durand S, Chery A, Pietrocola F, Vacchelli E, et al. Metabolomic analyses reveal that anti-aging metabolites are depleted by palmitate but increased by oleate in vivo. Cell Cycle. 2015;14:2399–407.
Bravo-San Pedro JM, Pietrocola F, Sica V, Izzo V, Sauvat A, Kepp O, et al. High-throughput quantification of GFP-LC3(+) dots by automated fluorescence microscopy. Methods in enzymology. 2017;587:71–86.
Linders PT, Horst CV, Beest MT, van den Bogaart G. Stx5-Mediated ER-Golgi transport in mammals and yeast. Cells. 2019;8.
Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–33.
Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, Scheuner D, et al. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23:169–79.
Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol cell. 2010;40:280–93.
Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016;17:1374–95.
Heal R, McGivan J. Induction of calreticulin expression in response to amino acid deprivation in Chinese hamster ovary cells. Biochem J. 1998;329(Pt 2):389–94.
Humeau J, Sauvat A, Cerrato G, Xie W, Loos F, Iannantuoni F, et al. Inhibition of transcription by dactinomycin reveals a new characteristic of immunogenic cell stress. EMBO Mol Med. 2020;12:e11622.
Taniuchi S, Miyake M, Tsugawa K, Oyadomari M, Oyadomari S. Integrated stress response of vertebrates is regulated by four eIF2alpha kinases. Sci Rep. 2016;6:32886.
Sauvat A, Leduc M, Muller K, Kepp O, Kroemer G. ColocalizR: An open-source application for cell-based high-throughput colocalization analysis. Comp Biol Med. 2019;107:227–34.
Lafay F. Envelope proteins of vesicular stomatitis virus: effect of temperature-sensitive mutations in complementation groups III and V. J Virol. 1974;14:1220–8.
Coria AS, Masseroni ML, Diaz, Anel AM. Regulation of PKD1-mediated Golgi to cell surface trafficking by Galphaq subunits. Biol Cell. 2014;106:30–43.
Boncompain G, Divoux S, Gareil N, de Forges H, Lescure A, Latreche L, et al. Synchronization of secretory protein traffic in populations of cells. Nat Meth. 2012;9:493–8.
Loos F, Xie W, Sica V, Bravo-San Pedro JM, Souquere S, Pierron G, et al. Artificial tethering of LC3 or p62 to organelles is not sufficient to trigger autophagy. Cell Death Dis. 2019;10:771.
Zhao L, Liu P, Boncompain G, Loos F, Lachkar S, Bezu L, et al. Identification of pharmacological inhibitors of conventional protein secretion. Sci Rep. 2018;8:14966.
Zhou H, Sauvat A, Gomes-da-Silva LC, Durand S, Forveille S, Iribarren K, et al. The oncolytic compound LTX-401 targets the Golgi apparatus. Cell Death Differ. 2018;25:227–8.
Talloczy Z, Jiang W, Virgin HWt, Leib DA, Scheuner D, Kaufman RJ, et al. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Nat Acad Sci USA. 2002;99:190–5.
Shen S, Niso-Santano M, Adjemian S, Takehara T, Malik SA, Minoux H, et al. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell. 2012;48:667–80.
Humeau J, Leduc M, Cerrato G, Loos F, Kepp O, Kroemer G. Phosphorylation of eukaryotic initiation factor-2α (eIF2α) in autophagy. 2020;11:433.
Geatti O, Shapiro B, Barillari B. Scintigraphic depiction of an insulinoma by I-131 metaiodobenzylguanidine. Clinical nuclear medicine. 1989;14:903–5.
Ha KD, Clarke BA, Brown WJ. Regulation of the Golgi complex by phospholipid remodeling enzymes. Biochimica et biophysica acta. 2012;1821:1078–88.
Casares D, Escriba PV, Rossello CA. Membrane lipid composition: effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. Int J Mol Sci. 2019;20.
Glaumann H, Motakefi AM, Jansson H. Intracellular distribution and effect of the antimalarial drug mefloquine on lysosomes of rat liver. Liver. 1992;12(4 Pt 1):183–90.
Parks A, Charest-Morin X, Boivin-Welch M, Bouthillier J, Marceau F. Autophagic flux inhibition and lysosomogenesis ensuing cellular capture and retention of the cationic drug quinacrine in murine models. PeerJ. 2015;3:e1314.
Hamaguchi M, Suzuki K, Fujita H, Uzuka T, Matsuda H, Shishido-Hara Y, et al. Successful treatment of non-HIV progressive multifocal leukoencephalopathy: case report and literature review. J Neurol. 2020;267:731–8.
Ugarte A, Danza A, Ruiz-Irastorza G. Glucocorticoids and antimalarials in systemic lupus erythematosus: an update and future directions. Current Opinion Rheumatol. 2018;30:482–9.
Ram A, Mabalirajan U, Singh SK, Singh VP, Ghosh B. Mepacrine alleviates airway hyperresponsiveness and airway inflammation in a mouse model of asthma. Int Immunopharmacol. 2008;8:893–9.
Alves P, Bashir MM, Wysocka M, Zeidi M, Feng R, Werth VP. Quinacrine suppresses tumor necrosis factor-alpha and IFN-alpha in Dermatomyositis and cutaneous lupus erythematosus. J Invest Dermatol Symp Proc. 2017;18:S57–S63.
Thase ME, Denko T. Pharmacotherapy of mood disorders. Annual Rev Clin Psych. 2008;4:53–91.
Cipriani A, Furukawa TA, Salanti G, Chaimani A, Atkinson LZ, Ogawa Y, et al. Comparative efficacy and acceptability of 21 antidepressant drugs for the acute treatment of adults with major depressive disorder: a systematic review and network meta-analysis. Lancet. 2018;391:1357–66.
Martins I, Kepp O, Schlemmer F, Adjemian S, Tailler M, Shen S, et al. Restoration of the immunogenicity of cisplatin-induced cancer cell death by endoplasmic reticulum stress. Oncogene. 2011;30:1147–58.
Chen G, Xie W, Nah J, Sauvat A, Liu P, Pietrocola F, et al. 3,4-Dimethoxychalcone induces autophagy through activation of the transcription factors TFE3 and TFEB. EMBO Mol Med. 2019;11:e10469.
Sauvat A, Cerrato G. Whole Genome Screenings results. figshare. Dataset. 2020. https://doi.org/10.6084/m9.figshare.13227686.
Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, et al. PubChem’s BioAssay Database. Nucleic Acids Res. 2012;40:D400–12.
Liu P, Zhao L, Loos F, Marty C, Xie W, Martins I, et al. Immunosuppression by mutated calreticulin released from malignant cells. Mol Cell. 2020;77:748–60 e9.
Acknowledgements
GC is supported by a scholarship of the Fondation pour la Recherche Médicale (FRM FDT202001011060). GK is supported by the Ligue contre le Cancer (équipe labellisée); Agence National de la Recherche (ANR)—Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; Chancellerie des universités de Paris (Legs Poix), Fondation pour la Recherche Médicale (FRM); a donation by Elior; European Research Area Network on Cardiovascular Diseases (ERA-CVD, MINOTAUR); Gustave Roussy Odyssea, the European Union Horizon 2020 Project Oncobiome; Fondation Carrefour; High-end Foreign Expert Program in China (GDW20171100085 and GDW20181100051), Institut National du Cancer (INCa); Inserm (HTE); Institut Universitaire de France; LeDucq Foundation; the LabEx Immuno-Oncology; the RHU Torino Lumière; the Seerave Foundation; the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); and the SIRIC Cancer Research and Personalized Medicine (CARPEM).
Author information
Authors and Affiliations
Contributions
Conceptualization, GK, AS, and GC; Methodology, GK, AS, GC, JH, ML, and PL; Investigation, GC, KM, PL, LZ, WX, and SZ; Formal analysis, AS, GC, and ML; Writing—Original Draft, GK, GC, and AS; Writing—Review & Editing, GK, GC, OK, and JH; Funding acquisition, GK; Supervision, OK and GK.
Corresponding authors
Ethics declarations
Conflict of interest
GK and OK are cofounders of Samsara Therapeutics. GK is a founder of everImmune and Therafast Bio.
Ethics
Animal experiments were conducted in compliance with the EU Directive 63/2010/EU from the European Parliament and were approved by the local Ethics Committee of the Gustave Roussy Cancer Campus (CEEA IRCIV/IGR no. 26, registered at the French Ministry of Research.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Piacentini
Supplementary information
Rights and permissions
About this article
Cite this article
Cerrato, G., Leduc, M., Müller, K. et al. Oleate-induced aggregation of LC3 at the trans-Golgi network is linked to a protein trafficking blockade. Cell Death Differ 28, 1733–1752 (2021). https://doi.org/10.1038/s41418-020-00699-3
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-020-00699-3
This article is cited by
-
Non-autophagic Golgi-LC3 lipidation facilitates TFE3 stress response against Golgi dysfunction
The EMBO Journal (2024)
-
A non-canonical role of ATG8 in Golgi recovery from heat stress in plants
Nature Plants (2023)
-
Everolimus and plicamycin specifically target chemoresistant colorectal cancer cells of the CMS4 subtype
Cell Death & Disease (2021)