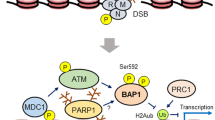
Abstract
The BAP1 gene has emerged as a major tumor suppressor mutated with various frequencies in numerous human malignancies, including uveal melanoma, malignant pleural mesothelioma, clear cell renal cell carcinoma, intrahepatic cholangiocarcinoma, hepatocellular carcinoma, and thymic epithelial tumors. BAP1 mutations are also observed at low frequency in other malignancies including breast, colorectal, pancreatic, and bladder cancers. BAP1 germline mutations are associated with high incidence of mesothelioma, uveal melanoma, and other cancers, defining the “BAP1 cancer syndrome.” Interestingly, germline BAP1 mutations constitute an important paradigm for gene–environment interactions, as loss of BAP1 predisposes to carcinogen-induced tumorigenesis. Inactivating mutations of BAP1 are also identified in sporadic cancers, denoting the importance of this gene for normal tissue homeostasis and tumor suppression, although some oncogenic properties have also been attributed to BAP1. BAP1 belongs to the deubiquitinase superfamily of enzymes, which are responsible for the maturation and turnover of ubiquitin as well as the reversal of substrate ubiquitination, thus regulating ubiquitin signaling. BAP1 is predominantly nuclear and interacts with several chromatin-associated factors, assembling multi-protein complexes with mutually exclusive partners. BAP1 exerts its function through highly regulated deubiquitination of its substrates. As such, BAP1 orchestrates chromatin-associated processes including gene expression, DNA replication, and DNA repair. BAP1 also exerts cytoplasmic functions, notably in regulating Ca2+ signaling at the endoplasmic reticulum. This DUB is also subjected to multiple post-translational modifications, notably phosphorylation and ubiquitination, indicating that several signaling pathways tightly regulate its function. Recent progress indicated that BAP1 plays essential roles in multiple cellular processes including cell proliferation and differentiation, cell metabolism, as well as cell survival and death. In this review, we summarize the biological and molecular functions of BAP1 and explain how the inactivation of this DUB might cause human cancers. We also highlight some of the unresolved questions and suggest potential new directions.

Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–112.
Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–7.
Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403.
Misaghi S, Ottosen S, Izrael-Tomasevic A, Arnott D, Lamkanfi M, Lee J, et al. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol. 2009;29:2181–92.
Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem. 2009;284:34179–88.
Yu H, Mashtalir N, Daou S, Hammond-Martel I, Ross J, Sui G, et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol. 2010;30:5071–85.
Mashtalir N, Daou S, Barbour H, Sen NN, Gagnon J, Hammond-Martel I, et al. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol Cell. 2014;54:392–406.
Okino Y, Machida Y, Frankland-Searby S, Machida YJ. BRCA1-associated protein 1 (BAP1) deubiquitinase antagonizes the ubiquitin-mediated activation of FoxK2 target genes. J Biol Chem. 2015;290:1580–91.
Daou S, Hammond-Martel I, Mashtalir N, Barbour H, Gagnon J, Iannantuono NV, et al. The BAP1/ASXL2 histone H2A deubiquitinase complex regulates cell proliferation and is disrupted in cancer. J Biol Chem. 2015;290:28643–63.
Daou S, Barbour H, Ahmed O, Masclef L, Baril C, Sen Nkwe N, et al. Monoubiquitination of ASXLs controls the deubiquitinase activity of the tumor suppressor BAP1. Nat Commun. 2018;9:4385.
Sahtoe DD, van Dijk WJ, Ekkebus R, Ovaa H, Sixma TK. BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat Commun. 2016;7:10292.
Ji Z, Mohammed H, Webber A, Ridsdale J, Han N, Carroll JS, et al. The forkhead transcription factor FOXK2 acts as a chromatin targeting factor for the BAP1-containing histone deubiquitinase complex. Nucleic Acids Res. 2014;42:6232–42.
Bononi A, Giorgi C, Patergnani S, Larson D, Verbruggen K, Tanji M, et al. BAP1 regulates IP3R3-mediated Ca(2+) flux to mitochondria suppressing cell transformation. Nature. 2017;546:549–53.
Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3.
Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–5.
Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–9.
Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–3.
Goldstein AM. Germline BAP1 mutations and tumor susceptibility. Nat Genet. 2011;43:925–6.
Chan-On W, Nairismagi ML, Ong CK, Lim WK, Dima S, Pairojkul C, et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet. 2013;45:1474–8.
Kovac M, Navas C, Horswell S, Salm M, Bardella C, Rowan A, et al. Recurrent chromosomal gains and heterogeneous driver mutations characterise papillary renal cancer evolution. Nat Commun. 2015;6:6336.
Bueno R, Stawiski EW, Goldstein LD, Durinck S, De Rienzo A, Modrusan Z, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48:407–16.
Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–72.
Woo HG, Choi JH, Yoon S, Jee BA, Cho EJ, Lee JH, et al. Integrative analysis of genomic and epigenomic regulation of the transcriptome in liver cancer. Nat Commun. 2017;8:839.
Karlsson J, Nilsson LM, Mitra S, Alsen S, Shelke GV, Sah VR, et al. Molecular profiling of driver events in metastatic uveal melanoma. Nat Commun. 2020;11:1894.
Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, Ulz P, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–21.
Abdel-Rahman MH, Pilarski R, Cebulla CM, Massengill JB, Christopher BN, Boru G, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48:856–9.
Carbone M, Ferris LK, Baumann F, Napolitano A, Lum CA, Flores EG, et al. BAP1 cancer syndrome: malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J Transl Med. 2012;10:179.
Njauw CN, Kim I, Piris A, Gabree M, Taylor M, Lane AM, et al. Germline BAP1 inactivation is preferentially associated with metastatic ocular melanoma and cutaneous-ocular melanoma families. PLoS ONE. 2012;7:e35295.
Farley MN, Schmidt LS, Mester JL, Pena-Llopis S, Pavia-Jimenez A, Christie A, et al. A novel germline mutation in BAP1 predisposes to familial clear-cell renal cell carcinoma. Mol Cancer Res. 2013;11:1061–71.
Popova T, Hebert L, Jacquemin V, Gad S, Caux-Moncoutier V, Dubois-d’Enghien C, et al. Germline BAP1 mutations predispose to renal cell carcinomas. Am J Hum Genet. 2013;92:974–80.
Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. BAP1 and cancer. Nat Rev Cancer. 2013;13:153–9.
Carbone M, Harbour JW, Brugarolas J, Bononi A, Pagano I, Dey A, et al. Biological mechanisms and clinical significance of BAP1 mutations in human cancer. Cancer Discov. 2020;10:1103–20.
Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–6.
Wang SS, Gu YF, Wolff N, Stefanius K, Christie A, Dey A, et al. Bap1 is essential for kidney function and cooperates with Vhl in renal tumorigenesis. Proc Natl Acad Sci USA. 2014;111:16538–43.
Napolitano A, Pellegrini L, Dey A, Larson D, Tanji M, Flores EG, et al. Minimal asbestos exposure in germline BAP1 heterozygous mice is associated with deregulated inflammatory response and increased risk of mesothelioma. Oncogene. 2016;35:1996–2002.
Kadariya Y, Cheung M, Xu J, Pei J, Sementino E, Menges CW, et al. Bap1 is a bona fide tumor suppressor: genetic evidence from mouse models carrying heterozygous germline Bap1 mutations. Cancer Res. 2016;76:2836–44.
Gu YF, Cohn S, Christie A, McKenzie T, Wolff N, Do QN, et al. Modeling renal cell carcinoma in mice: Bap1 and Pbrm1 inactivation drive tumor grade. Cancer Discov. 2017;7:900–17.
Kukuyan AM, Sementino E, Kadariya Y, Menges CW, Cheung M, Tan Y, et al. Inactivation of Bap1 cooperates with losses of Nf2 and Cdkn2a to drive the development of pleural malignant mesothelioma in conditional mouse models. Cancer Res. 2019;79:4113–23.
Badhai J, Pandey GK, Song JY, Krijgsman O, Bhaskaran R, Chandrasekaran G, et al. Combined deletion of Bap1, Nf2, and Cdkn2ab causes rapid onset of malignant mesothelioma in mice. J Exp Med. 2020;217:e20191257.
Perkail S, Andricovich J, Kai Y, Tzatsos A. BAP1 is a haploinsufficient tumor suppressor linking chronic pancreatitis to pancreatic cancer in mice. Nat Commun. 2020;11:3018.
Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8.
de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–76.
Pengelly AR, Kalb R, Finkl K, Muller J. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev. 2015;29:1487–92.
Wang L, Zhao Z, Ozark PA, Fantini D, Marshall SA, Rendleman EJ, et al. Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nat Med. 2018;24:758–69.
Campagne A, Lee MK, Zielinski D, Michaud A, Le Corre S, Dingli F, et al. BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat Commun. 2019;10:348.
He M, Chaurushiya MS, Webster JD, Kummerfeld S, Reja R, Chaudhuri S, et al. Intrinsic apoptosis shapes the tumor spectrum linked to inactivation of the deubiquitinase BAP1. Science. 2019;364:283–5.
Kolovos P, Nishimura K, Sankar A, Sidoli S, Cloos PA, Helin K, et al. PR-DUB maintains expression of critical genes through FOXK1/2 and ASXL1/2/3-dependent recruitment to chromatin and H2AK119ub1 deubiquitination. Genome Res. 2020;30:1119–30.
Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911.
Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–49.
Narayanan A, Ruyechan WT, Kristie TM. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci USA. 2007;104:10835–40.
Julien E, Herr W. A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol Cell. 2004;14:713–25.
Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–19.
Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med. 2009;15:1312–7.
Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–12.
Peng H, Nogueira ML, Vogel JL, Kristie TM. Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components. Proc Natl Acad Sci USA. 2010;107:2461–6.
Revenko AS, Kalashnikova EV, Gemo AT, Zou JX, Chen HW. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol Cell Biol. 2010;30:5260–72.
Yu L, Jearawiriyapaisarn N, Lee MP, Hosoya T, Wu Q, Myers G, et al. BAP1 regulation of the key adaptor protein NCoR1 is critical for gamma-globin gene repression. Genes Dev. 2018;32:1537–49.
Dai F, Lee H, Zhang Y, Zhuang L, Yao H, Xi Y, et al. BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proc Natl Acad Sci USA. 2017;114:3192–7.
Zhang Y, Shi J, Liu X, Feng L, Gong Z, Koppula P, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol. 2018;20:1181–92.
Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807.
Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46:722–34.
Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204.
van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–17.
Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–45.
Yu H, Pak H, Hammond-Martel I, Ghram M, Rodrigue A, Daou S, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci USA. 2014;111:285–90.
Ismail IH, Davidson R, Gagne JP, Xu ZZ, Poirier GG, Hendzel MJ. Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res. 2014;74:4282–94.
Lee HS, Lee SA, Hur SK, Seo JW, Kwon J. Stabilization and targeting of INO80 to replication forks by BAP1 during normal DNA synthesis. Nat Commun. 2014;5:5128.
Lee HS, Seo HR, Lee SA, Choi S, Kang D, Kwon J. BAP1 promotes stalled fork restart and cell survival via INO80 in response to replication stress. Biochem J. 2019;476:3053–66.
Watanabe S, Peterson CL. The INO80 family of chromatin-remodeling enzymes: regulators of histone variant dynamics. Cold Spring Harb Symp Quant Biol. 2010;75:35–42.
Gerhold CB, Gasser SM. INO80 and SWR complexes: relating structure to function in chromatin remodeling. Trends Cell Biol. 2014;24:619–31.
Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008;68:6953–62.
Arenzana TL, Lianoglou S, Seki A, Eidenschenk C, Cheung T, Seshasayee D, et al. Tumor suppressor BAP1 is essential for thymic development and proliferative responses of T lymphocytes. Sci Immunol. 2018;3:eaal1953.
Pan H, Jia R, Zhang L, Xu S, Wu Q, Song X, et al. BAP1 regulates cell cycle progression through E2F1 target genes and mediates transcriptional silencing via H2A monoubiquitination in uveal melanoma cells. Int J Biochem Cell Biol. 2015;60:176–84.
van Ginkel PR, Hsiao KM, Schjerven H, Farnham PJ. E2F-mediated growth regulation requires transcription factor cooperation. J Biol Chem. 1997;272:18367–74.
Schlisio S, Halperin T, Vidal M, Nevins JR. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–86.
Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, et al. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol. 2006;26:3565–81.
Xu J, Kadariya Y, Cheung M, Pei J, Talarchek J, Sementino E, et al. Germline mutation of Bap1 accelerates development of asbestos-induced malignant mesothelioma. Cancer Res. 2014;74:4388–97.
Qin J, Zhou Z, Chen W, Wang C, Zhang H, Ge G, et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471.
Chen XX, Yin Y, Cheng JW, Huang A, Hu B, Zhang X, et al. BAP1 acts as a tumor suppressor in intrahepatic cholangiocarcinoma by modulating the ERK1/2 and JNK/c-Jun pathways. Cell Death Dis. 2018;9:1036.
Lee HJ, Pham T, Chang MT, Barnes D, Cai AG, Noubade R, et al. The tumor suppressor BAP1 regulates the hippo pathway in pancreatic ductal adenocarcinoma. Cancer Res. 2020;80:1656–68.
Deng R, Guo Y, Li L, He J, Qiang Z, Zhang H, et al. BAP1 suppresses prostate cancer progression by deubiquitinating and stabilizing PTEN. Mol Oncol. 2020;15:279–98.
Blanpain C, Daley GQ, Hochedlinger K, Passegue E, Rossant J, Yamanaka S. Stem cells assessed. Nat Rev Mol Cell Biol. 2012;13:471–6.
Laurenti E, Gottgens B. From haematopoietic stem cells to complex differentiation landscapes. Nature. 2018;553:418–26.
Heidstra R, Sabatini S. Plant and animal stem cells: similar yet different. Nat Rev Mol Cell Biol. 2014;15:301–12.
Goodell MA, Rando TA. Stem cells and healthy aging. Science. 2015;350:1199–204.
Wu J, Izpisua Belmonte JC. Stem cells: a renaissance in human biology research. Cell. 2016;165:1572–85.
Baughman JM, Rose CM, Kolumam G, Webster JD, Wilkerson EM, Merrill AE, et al. NeuCode proteomics reveals Bap1 regulation of metabolism. Cell Rep. 2016;16:583–95.
LaFave LM, Beguelin W, Koche R, Teater M, Spitzer B, Chramiec A, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21:1344–9.
Kuznetsov JN, Aguero TH, Owens DA, Kurtenbach S, Field MG, Durante MA, et al. BAP1 regulates epigenetic switch from pluripotency to differentiation in developmental lineages giving rise to BAP1-mutant cancers. Sci Adv. 2019;5:eaax1738.
Danese A, Patergnani S, Bonora M, Wieckowski MR, Previati M, Giorgi C, et al. Calcium regulates cell death in cancer: roles of the mitochondria and mitochondria-associated membranes (MAMs). Biochim Biophys Acta Bioenerg. 2017;1858:615–27.
Marchi S, Patergnani S, Missiroli S, Morciano G, Rimessi A, Wieckowski MR, et al. Mitochondrial and endoplasmic reticulum calcium homeostasis and cell death. Cell Calcium. 2018;69:62–72.
Stefan CJ. Endoplasmic reticulum-plasma membrane contacts: Principals of phosphoinositide and calcium signaling. Curr Opin Cell Biol. 2020;63:125–34.
Bhosale G, Sharpe JA, Sundier SY, Duchen MR. Calcium signaling as a mediator of cell energy demand and a trigger to cell death. Ann N Y Acad Sci. 2015;1350:107–16.
Bauer TM, Murphy E. Role of mitochondrial calcium and the permeability transition pore in regulating cell death. Circ Res. 2020;126:280–93.
Sime W, Niu Q, Abassi Y, Masoumi KC, Zarrizi R, Kohler JB, et al. BAP1 induces cell death via interaction with 14-3-3 in neuroblastoma. Cell Death Dis. 2018;9:458.
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72.
Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–85.
Affar EB, Carbone M. BAP1 regulates different mechanisms of cell death. Cell Death Dis. 2018;9:1151.
Lei P, Bai T, Sun Y. Mechanisms of ferroptosis and relations with regulated cell death: a review. Front Physiol. 2019;10:139.
Deng T, Lyon CJ, Bergin S, Caligiuri MA, Hsueh WA. Obesity, inflammation, and cancer. Annu Rev Pathol. 2016;11:421–49.
Goodpaster BH, Sparks LM. Metabolic flexibility in health and disease. Cell Metab. 2017;25:1027–36.
Bantug GR, Galluzzi L, Kroemer G, Hess C. The spectrum of T cell metabolism in health and disease. Nat Rev Immunol. 2018;18:19–34.
Liu G, Summer R. Cellular metabolism in lung health and disease. Annu Rev Physiol. 2019;81:403–28.
Bononi A, Yang H, Giorgi C, Patergnani S, Pellegrini L, Su M, et al. Germline BAP1 mutations induce a Warburg effect. Cell Death Differ. 2017;24:1694–704.
Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S, et al. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. Cell Metab. 2012;16:226–37.
Izawa T, Rohatgi N, Fukunaga T, Wang QT, Silva MJ, Gardner MJ, et al. ASXL2 regulates glucose, lipid, and skeletal homeostasis. Cell Rep. 2015;11:1625–37.
Yang C, Ding H, Yang Y, Yang L, Yang Y, Fang M, et al. BAP1 regulates AMPK-mTOR signalling pathway through deubiquitinating and stabilizing tumour-suppressor LKB1. Biochem Biophys Res Commun. 2020;529:1025–32.
Hauri S, Comoglio F, Seimiya M, Gerstung M, Glatter T, Hansen K, et al. A high-density map for navigating the human polycomb complexome. Cell Rep. 2016;17:583–95.
Daou S, Mashtalir N, Hammond-Martel I, Pak H, Yu H, Sui G, et al. Crosstalk between O-GlcNAcylation and proteolytic cleavage regulates the host cell factor-1 maturation pathway. Proc Natl Acad Sci U S A. 2011;108:2747–52.
Capotosti F, Guernier S, Lammers F, Waridel P, Cai Y, Jin J, et al. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 2011;144:376–88.
Yang Y, Yin X, Yang H, Xu Y. Histone demethylase LSD2 acts as an E3 ubiquitin ligase and inhibits cancer cell growth through promoting proteasomal degradation of OGT. Mol Cell. 2015;58:47–59.
Xiong Z, Xia P, Zhu X, Geng J, Wang S, Ye B, et al. Glutamylation of deubiquitinase BAP1 controls self-renewal of hematopoietic stem cells and hematopoiesis. J Exp Med. 2020;217:e20190974.
Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–62.
Rogowski K, van Dijk J, Magiera MM, Bosc C, Deloulme JC, Bosson A, et al. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 2010;143:564–78.
Field MG, Durante MA, Anbunathan H, Cai LZ, Decatur CL, Bowcock AM, et al. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat Commun. 2018;9:116.
Brouwer NJ, Gezgin G, Wierenga APA, Bronkhorst IHG, Marinkovic M, Luyten GPM, et al. Tumour angiogenesis in uveal melanoma is related to genetic evolution. Cancers. 2019;11:979.
Yoshikawa Y, Emi M, Hashimoto-Tamaoki T, Ohmuraya M, Sato A, Tsujimura T, et al. High-density array-CGH with targeted NGS unmask multiple noncontiguous minute deletions on chromosome 3p21 in mesothelioma. Proc Natl Acad Sci USA. 2016;113:13432–7.
Hmeljak J, Sanchez-Vega F, Hoadley KA, Shih J, Stewart C, Heiman D, et al. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov. 2018;8:1548–65.
Shrestha R, Nabavi N, Lin YY, Mo F, Anderson S, Volik S, et al. BAP1 haploinsufficiency predicts a distinct immunogenic class of malignant peritoneal mesothelioma. Genome Med. 2019;11:8.
Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–33.
Liao L, Liu ZZ, Langbein L, Cai W, Cho EA, Na J, et al. Multiple tumor suppressors regulate a HIF-dependent negative feedback loop via ISGF3 in human clear cell renal cancer. Elife. 2018;7:e37925.
Huang Y, Wang J, Jia P, Li X, Pei G, Wang C, et al. Clonal architectures predict clinical outcome in clear cell renal cell carcinoma. Nat Commun. 2019;10:1245.
Chen YB, Xu J, Skanderup AJ, Dong Y, Brannon AR, Wang L, et al. Molecular analysis of aggressive renal cell carcinoma with unclassified histology reveals distinct subsets. Nat Commun. 2016;7:13131.
Mosbeh A, Halfawy K, Abdel-Mageed WS, Sweed D, Rahman MHA. Nuclear BAP1 loss is common in intrahepatic cholangiocarcinoma and a subtype of hepatocellular carcinoma but rare in pancreatic ductal adenocarcinoma. Cancer Genet. 2018;224-5:21–8.
Wang Y, Thomas A, Lau C, Rajan A, Zhu Y, Killian JK, et al. Mutations of epigenetic regulatory genes are common in thymic carcinomas. Sci Rep. 2014;4:7336.
Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4.
Je EM, Lee SH, Yoo NJ. Somatic mutation of a tumor suppressor gene BAP1 is rare in breast, prostate, gastric and colorectal cancers. APMIS. 2012;120:855–6.
Tayao M, Andrici J, Farzin M, Clarkson A, Sioson L, Watson N, et al. Loss of BAP1 expression is very rare in pancreatic ductal adenocarcinoma. PLoS ONE. 2016;11:e0150338.
Tesch ME, Pater JA, Vandekerkhove G, Wang G, Binnington K, So AI, et al. Concurrent germline and somatic pathogenic BAP1 variants in a patient with metastatic bladder cancer. NPJ Genom Med. 2020;5:12.
Carbone M, Flores EG, Emi M, Johnson TA, Tsunoda T, Behner D, et al. Combined genetic and genealogic studies uncover a large BAP1 cancer syndrome kindred tracing back nine generations to a common ancestor from the 1700s. PLoS Genet. 2015;11:e1005633.
Walpole S, Pritchard AL, Cebulla CM, Pilarski R, Stautberg M, Davidorf FH, et al. Comprehensive study of the clinical phenotype of germline BAP1 variant-carrying families worldwide. J Natl Cancer Inst. 2018;110:1328–41.
Ohar JA, Cheung M, Talarchek J, Howard SE, Howard TD, Hesdorffer M, et al. Germline BAP1 mutational landscape of asbestos-exposed malignant mesothelioma patients with family history of cancer. Cancer Res. 2016;76:206–15.
Rawson RV, Watson GF, Maher AM, McCarthy SW, Thompson JF, Scolyer RA. Germline BAP1 mutations also predispose to cutaneous squamous cell carcinoma. Pathology. 2017;49:539–42.
Bhattacharya S, Hanpude P, Maiti TK. Cancer associated missense mutations in BAP1 catalytic domain induce amyloidogenic aggregation: a new insight in enzymatic inactivation. Sci Rep. 2015;5:18462.
Guo G, Chmielecki J, Goparaju C, Heguy A, Dolgalev I, Carbone M, et al. Whole-exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer Res. 2015;75:264–9.
Farquhar N, Thornton S, Coupland SE, Coulson JM, Sacco JJ, Krishna Y, et al. Patterns of BAP1 protein expression provide insights into prognostic significance and the biology of uveal melanoma. J Pathol Clin Res. 2018;4:26–38.
Asada S, Goyama S, Inoue D, Shikata S, Takeda R, Fukushima T, et al. Mutant ASXL1 cooperates with BAP1 to promote myeloid leukaemogenesis. Nat Commun. 2018;9:2733.
Steurer S, Schwemmer L, Hube-Magg C, Buscheck F, Hoflmayer D, Tsourlakis MC, et al. Nuclear up regulation of the BRCA1-associated ubiquitinase BAP1 is associated with tumor aggressiveness in prostate cancers lacking the TMPRSS2:ERG fusion. Oncotarget. 2019;10:7096–111.
Gelsi-Boyer V, Trouplin V, Adelaide J, Bonansea J, Cervera N, Carbuccia N, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145:788–800.
Boultwood J, Perry J, Pellagatti A, Fernandez-Mercado M, Fernandez-Santamaria C, Calasanz MJ, et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010;24:1062–5.
Fisher CL, Pineault N, Brookes C, Helgason CD, Ohta H, Bodner C, et al. Loss-of-function additional sex combs like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia. Blood. 2010;115:38–46.
Gelsi-Boyer V, Trouplin V, Roquain J, Adelaide J, Carbuccia N, Esterni B, et al. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br J Haematol. 2010;151:365–75.
Katoh M. Functional and cancer genomics of ASXL family members. Br J Cancer. 2013;109:299–306.
Micol JB, Abdel-Wahab O. The role of additional sex combs-like proteins in cancer. Cold Spring Harb Perspect Med. 2016;6:a026526.
Balasubramani A, Larjo A, Bassein JA, Chang X, Hastie RB, Togher SM, et al. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat Commun. 2015;6:7307.
Yang H, Kurtenbach S, Guo Y, Lohse I, Durante MA, Li J, et al. Gain of function of ASXL1 truncating protein in the pathogenesis of myeloid malignancies. Blood. 2018;131:328–41.
Nagase R, Inoue D, Pastore A, Fujino T, Hou HA, Yamasaki N, et al. Expression of mutant Asxl1 perturbs hematopoiesis and promotes susceptibility to leukemic transformation. J Exp Med. 2018;215:1729–47.
Kolluri KK, Alifrangis C, Kumar N, Ishii Y, Price S, Michaut M, et al. Loss of functional BAP1 augments sensitivity to TRAIL in cancer cells. Elife. 2018;7:e30224.
Ladanyi M, Sanchez Vega F, Zauderer M. Loss of BAP1 as a candidate predictive biomarker for immunotherapy of mesothelioma. Genome Med. 2019;11:18.
Figueiredo CR, Kalirai H, Sacco JJ, Azevedo RA, Duckworth A, Slupsky JR, et al. Loss of BAP1 expression is associated with an immunosuppressive microenvironment in uveal melanoma, with implications for immunotherapy development. J Pathol. 2020;250:420–39.
Acknowledgements
This work was supported by grants from the Canadian Institutes of Health Research (2018–2023) to EBA. EBA is a senior scholar of the Fonds de la Recherche du Québec-Santé (FRQ-S). LM holds a PhD scholarship from the FRQ-S. AO holds PhD scholarships from the FRQ-S and the Cole Foundation. Certain forms and shapes displayed in figures were taken and modified from Servier Medical Art, licensed under a Creative Common Attribution 3.0 Generic License. http://smart.servier.com/.
Author information
Authors and Affiliations
Contributions
EBA, ML, AO, and BE collected the references and wrote the manuscript. ML, AO, BE, and EBA realized the illustration. BL, NL, and AN contributed to the general improvement of the writing and critical review of the figures.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by F. Pentimalli
Rights and permissions
About this article
Cite this article
Masclef, L., Ahmed, O., Estavoyer, B. et al. Roles and mechanisms of BAP1 deubiquitinase in tumor suppression. Cell Death Differ 28, 606–625 (2021). https://doi.org/10.1038/s41418-020-00709-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-020-00709-4
This article is cited by
-
Regulation of ferroptosis by BAP1
Cell Death & Differentiation (2026)
-
Restoration of BAP1 activity via base editing suppresses anchorage-independent survival in kidney cancer
Cancer Cell International (2025)
-
AMPK-activated BAP1 regulates pVHL stability and tumor-suppressive functions
Cell Death & Differentiation (2025)
-
Pleural mesothelioma
Nature Reviews Disease Primers (2025)
-
Epigenetic regulation of ACSL4 via H2A monoubiquitylation connects lipid metabolism to BAP1-mediated ferroptosis
Cell Death & Differentiation (2025)