Abstract
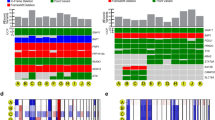
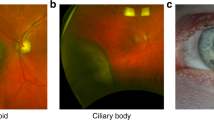
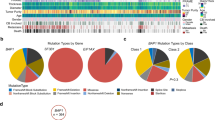
Intratumor heterogeneity has been recognized in numerous cancers as a major source of metastatic dissemination. In uveal melanomas, the existence and identity of specific subpopulations, their biological function and their contribution to metastasis remain unknown. Here, in multiscale analyses using single-cell RNA sequencing of six different primary uveal melanomas, we uncover an intratumoral heterogeneity at the genomic and transcriptomic level. We identify distinct transcriptional cell states and diverse tumor-associated populations in a subset of the samples. We also decipher a gene regulatory network underlying an invasive and poor prognosis state driven in part by the transcription factor HES6. HES6 heterogenous expression has been validated by RNAscope assays within primary human uveal melanomas, which further unveils the existence of these cells conveying a dismal prognosis in tumors diagnosed with a favorable outcome using bulk analyses. Depletion of HES6 impairs proliferation, migration and metastatic dissemination in vitro and in vivo using the chick chorioallantoic membrane assay, demonstrating the essential role of HES6 in uveal melanomas. Thus, single-cell analysis offers an unprecedented view of primary uveal melanoma heterogeneity, identifies bona fide biomarkers for metastatic cells in the primary tumor, and reveals targetable modules driving growth and metastasis formation. Significantly, our findings demonstrate that HES6 is a valid target to stop uveal melanoma progression.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
The experimental data from single-cell RNA sequencing, whole exome sequencing and array-CGH have been deposited in the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) under the SuperSeries GSE138665.
References
Rietschel P, Panageas KS, Hanlon C, Patel A, Abramson DH, Chapman PB. Variates of survival in metastatic uveal melanoma. J Clin Oncol. 2005;23:8076–80.
Nabil AA, Marie S, Marc-Henri S, Nathalie C, Laurence D, Sophie PN, et al. Upcoming translational challenges for uveal melanoma. Br J Cancer. 2015;113:1249–53.
Pandiani C, Beranger GE, Leclerc J, Ballotti R, Bertolotto C. Focus on cutaneous and uveal melanoma specificities. Genes Dev. 2017;31:724–43.
Yang J, Manson DK, Marr BP, Carvajal RD. Treatment of uveal melanoma: where are we now? Ther Adv Med Oncol. 2018. https://doi.org/10.1177/1758834018757175.
Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–96.
Kemper K, Krijgsman O, Cornelissen-Steijger P, Shahrabi A, Weeber F, Song JY, et al. Intra- and inter-tumor heterogeneity in a vemurafenib-resistant melanoma patient and derived xenografts. EMBO Mol Med. 2015;7:1104–18.
Rambow F, Marine J, Goding CR. Melanoma plasticity and phenotypic diversity: therapeutic barriers and opportunities. Genes Dev. 2019;33:1295–318.
Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–9.
Durante MA, Rodriguez DA, Kurtenbach S, Kuznetsov JN, Sanchez MI, Decatur CL, et al. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat Commun. 2020;11:496.
Karlsson J, Nilsson LM, Mitra S, Alsén S, Shelke GV, Sah VR, et al. Molecular profiling of driver events in metastatic uveal melanoma. Nat Commun. 2020;11:1894.
Figueiredo CR, Kalirai H, Sacco JJ, Azevedo RA, Duckworth A, Slupsky JR, et al. Loss of BAP1 expression is associated with an immunosuppressive microenvironment in uveal melanoma, with implications for immunotherapy development. J Pathol. 2020;250:420–39.
Triozzi PL, Schoenfield L, Plesec T, Saunthararajah Y, Tubbs RR, Singh AD. Molecular profiling of primary uveal melanomas with tumor-infiltrating lymphocytes. Oncoimmunology. 2014;8:e947169.
Chen PW, Murray TG, Uno T, Salgaller ML, Reddy RKB. Expression of MAGE genes in ocular melanoma during progression from primary to metastatic disease. Clin Exp Metastasis. 1997;15:509–18.
De Waard‐Siebinga I, Blom DR, Griffioen M, Schrier PI, Hoogendoorn E, Beverstock G, et al. Establishment and characterization of an uveal‐melanoma cell line. Int J Cancer. 1995;62:155–61.
Luyten GP, Naus NC, Mooy CM, Hagemeijer A, Kan-Mitchell J, Van Drunen E, et al. Establishment and characterization of primary and metastatic uveal melanoma cell lines. Int J Cancer. 1996;66:380–7.
Griewank KG, Yu X, Khalili J, Sozen MM, Stempke-Hale K, Bernatchez C, et al. Genetic and molecular characterization of uveal melanoma cell lines. Pigment Cell Melanoma Res. 2012;25:182–7.
Shain AH, Bagger MM, Yu R, Chang D, Liu S, Vemula S, et al. The genetic evolution of metastatic uveal melanoma. Nat Genet. 2019;51:1123–30.
Larribère L, Utikal J. Update on gna alterations in cancer: Implications for uveal melanoma treatment. Cancers. 2020;12:1–18.
Johansson P, Aoude LG, Wadt K, Glasson WJ, Warrier SK, Hewitt AW, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7:4624–31.
Moore AR, Ceraudo E, Sher JJ, Guan Y, Shoushtari AN, Chang MT, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet. 2016;48:675–80.
Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW, et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32:4814–24.
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–401.
Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–6.
Meir T, Dror R, Yu X, Qian J, Simon I, Pe’er J, et al. Molecular characteristics of liver metastases from uveal melanoma. Invest Ophthalmol Vis Sci. 2007;48:4890–6.
Xu Y, Liu X, Zhang H, Zhu Z, Wu X, Wu X, et al. Overexpression of HES6 has prognostic value and promotes metastasis via the Wnt/beta-catenin signaling pathway in colorectal cancer. Oncol Rep. 2018;40:1261–74.
Haapa-Paananen S, Kiviluoto S, Waltari M, Puputti M, Mpindi JP, Kohonen P, et al. HES6 gene is selectively overexpressed in glioma and represents an important transcriptional regulator of glioma proliferation. Oncogene. 2012;31:1299–310.
Wickramasinghe CM, Domaschenz R, Amagase Y, Williamson D, Missiaglia E, Shipley J, et al. HES6 enhances the motility of alveolar rhabdomyosarcoma cells. Exp Cell Res. 2013;319:103–12.
Asnaghi L, Ebrahimi KB, Schreck KC, Bar EE, Coonfield ML, Bell WR, et al. Notch signaling promotes growth and invasion in uveal melanoma. Clin Cancer Res. 2012;18:654–65.
Asnaghi L, Handa JT, Merbs SL, Harbour JW, Eberhart CG. A role for Jag2 in promoting uveal melanoma dissemination and growth. Investig Ophthalmol Vis Sci. 2013;54:295–306.
Dopierala J, Damato BE, Lake SL, Taktak AF, Coupland SE. Genetic heterogeneity in uveal melanoma assessed by multiplex ligation-dependent probe amplification. Invest Ophthalmol Vis Sci. 2010;51:4898–905.
Goding CR, Arnheiter H. MITF—the first 25 years. Genes Dev. 2019;33:983–1007.
Cheli Y, Ohanna M, Ballotti R, Bertolotto C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res. 2010;23:27–40.
Ohanna M, Giuliano S, Bonet C, Imbert V, Hofman V, Zangari J, et al. Senescent cells develop a parp-1 and nuclear factor-κB-associated secretome (PNAS). Genes Dev. 2011;25:1245–61.
Ohanna M, Cheli Y, Bonet C, Bonazzi VF, Allegra M, Giuliano S, et al. Secretome from senescent melanoma engages the STAT3 pathway to favor reprogramming of naive melanoma towards a tumor-initiating cell phenotype. Oncotarget. 2013;4:2212–24.
Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–31.
Chang TP, Vancurova I. Bcl3 regulates pro-survival and pro-inflammatory gene expression in cutaneous T-cell lymphoma. Biochim Biophys Acta - Mol Cell Res. 2014;1843:2620–30.
Riesenberg S, Groetchen A, Siddaway R, Bald T, Reinhardt J, Smorra D, et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat Commun. 2015;6:8755.
Landsberg J, Kohlmeyer J, Renn M, Bald T, Rogava M, Cron M, et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490:412–6.
Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204–20 e15.
Lawson CD, Burridge K. The on-off relationship of Rho and Rac during integrin-mediated adhesion and cell migration. Small GTPases. 2014;5:e27958.
Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell. 2014;25:831–45.
Miskolczi Z, Smith MP, Rowling EJ, Ferguson J, Barriuso J, Wellbrock C. Collagen abundance controls melanoma phenotypes through lineage-specific microenvironment sensing. Oncogene. 2018;37:3166–82.
Roma J, Masià A, Reventós J, De Toledo JS, Gallego S. Notch pathway inhibition significantly reduces rhabdomyosarcoma invasiveness and mobility in vitro. Clin Cancer Res. 2011;17:505–13.
Trolet J, Hupe P, Huon I, Lebigot I, Decraene C, Delattre O, et al. Genomic profiling and identification of high-risk uveal melanoma by array CGH analysis of primary tumors and liver metastases. Invest Ophthalmol Vis Sci. 2009;50:2572–80.
Acknowledgements
We thank the Single Cell Core Facility at IPMC and the imaging facility of C3M. The authors thank all patients with primary uveal melanoma for participating as well as Frédéric Reinier for the bioinformatics analysis at the beginning of the project. We also thank Dr. M. Jager (Leiden, The Netherlands) for providing the primary uveal melanoma cell lines.
Funding
This work was supported by the French government, INSERM, La Ligue Nationale contre le cancer, INCA PLBio to CB and PB (INCA-12824), Conseil Départemental des Alpes Maritimes (2016-294DGADSH-CV), the National Research Agency under the Investments for the Futur program UCAJEDI «ANR-15-IDEX-01», National Infrastructure France Génomique «ANR-10-INBS-09-03» and «ANR-10-INBS-09-02», Canceropole PACA, Club Francophone des spécialistes de la rétine (CFSR), La Ville de Nice, Mickaël Fulci, La Fondation ARC to CB (20171206312) and La Fondation pour la Recherche Médicale for CP and GG. ID is an ‘équipe labélisée’ of the French League against Cancer.
Author information
Authors and Affiliations
Contributions
CB and RB designed and supervised the study, prepared the figures and wrote the manuscript. VM carried out scRNA-seq experiments. KLB and NN performed the computational analyses and assisted in data analysis in discussions with PB who also critically reviewed the manuscript. CP, TS, YC and GB performed patient sample processing and functional experiments. KB, CH, MD and MI provided technical assistance. GG and ID carried out the RNAscope assays. SL and PH coordinated patient sample collection, maintained IRB approval and performed histological analysis. FP provided cytogenetic analyses. AM, SNE, CM, JPC and SB gathered patients’ consent, provided the samples and clinical data.
Corresponding author
Ethics declarations
Ethics
The study was approved by the hospital ethics committee (Nice Hospital Center and University of Nice Côte d’Azur). The study was performed in accordance with the Declaration of Helsinki.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Blagosklonny
Supplementary information
Rights and permissions
About this article
Cite this article
Pandiani, C., Strub, T., Nottet, N. et al. Single-cell RNA sequencing reveals intratumoral heterogeneity in primary uveal melanomas and identifies HES6 as a driver of the metastatic disease. Cell Death Differ 28, 1990–2000 (2021). https://doi.org/10.1038/s41418-020-00730-7
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-020-00730-7
This article is cited by
-
CanCellCap: robust cancer cell capture across tissue types on single-cell RNA-seq data by multi-domain learning
BMC Biology (2025)
-
Spatially resolved transcriptomics reveals gene expression characteristics in uveal melanoma
Holistic Integrative Oncology (2025)
-
Pan-cancer analysis reveals dual roles of HES6 in cancer immunity and prognosis
Discover Oncology (2025)
-
Integrating bulk and single-cell RNA sequencing reveals intratumor heterogeneity phenotypes and immune infiltration in pancreatic cancer
Discover Oncology (2025)
-
SETDB1 is critically required for uveal melanoma growth and represents a promising therapeutic target
Cell Death & Disease (2025)