Abstract
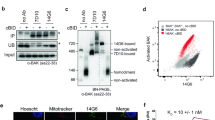
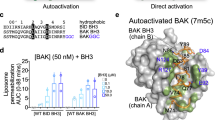
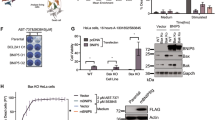
The intricate interplay among BCL-2 family proteins governs mitochondrial apoptosis, with the anti-apoptotic protein MCL-1 primarily exerting its function by sequestering the pore-forming effector BAK. Understanding the MCL-1/BAK complex is pivotal for the sensitivity of cancer cells to BH3 mimetics, yet the precise molecular mechanism underlying their interaction remains elusive. Herein, we demonstrate that a canonical BH3 peptide from BAK inadequately binds to MCL-1 proteins, whereas an extended BAK-BH3 peptide with five C-terminal residues exhibits a remarkable 65-fold increase in affinity. By elucidating the complex structures of MCL-1 bound to these two BAK-BH3 peptides at 2.08 Å and 1.98 Å resolutions, we uncover their distinct binding specificities. Notably, MCL-1 engages in critical hydrophobic interactions with the extended BAK-BH3 peptide, particularly at an additional p5 sub-pocket, featuring a π-π stacking interaction between MCL-1 Phe319 and BAK Tyr89. Mutations within this p5 sub-pocket substantially disrupt the MCL-1/BAK protein-protein interaction. Furthermore, the p5 sub-pocket of MCL-1 significantly influences the efficacy of MCL-1 inhibitors. Overall, our findings elucidate the molecular specificity underlying MCL-1 binding to BAK and underscore the significance of the p5 hydrophobic sub-pocket in their high-affinity interaction, thus providing novel insights for the development of BH3 mimetics targeting the MCL-1/BAK interaction as potential therapeutics for cancer treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The coordinates and structure factors are deposited in the Protein Data Bank under the accession codes 8Y1Y (MCL-1/BAK-BH3long structure), 8Y1Z (MCL-1/BAK-BH3short structure) and 8Y20 (MCL-1/A-1210477 structure). Other data generated in this study are provided in the article and in the Supplementary Materials.
References
Newton K, Strasser A, Kayagaki N, Dixit VM. Cell death. Cell. 2024;187:235–56.
Czabotar PE, Garcia-Saez AJ. Mechanisms of BCL-2 family proteins in mitochondrial apoptosis. Nat Rev Mol Cell Bio. 2023;24:732–48.
Vandenabeele P, Bultynck G, Savvides SN. Pore-forming proteins as drivers of membrane permeabilization in cell death pathways. Nat Rev Mol Cell Bio. 2023;24:312–33.
Vitale I, Pietrocola F, Guilbaud E, Aaronson SA, Abrams JM, Adam D, et al. Apoptotic cell death in disease-Current understanding of the NCCD 2023. Cell Death Differ. 2023;30:1097–154.
Moldoveanu T, Czabotar PE. BAX, BAK, and BOK: a coming of age for the BCL-2 family effector proteins. Cold Spring Harb Perspect Biol. 2020;12:a036319.
Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, et al. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell. 2011;44:517–31.
Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–305.
Hockings C, Alsop AE, Fennell SC, Lee EF, Fairlie WD, Dewson G, et al. Mcl-1 and Bcl-xL sequestration of Bak confers differential resistance to BH3-only proteins. Cell Death Differ. 2018;25:719–32.
Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–9.
Huang K, O’Neill KL, Li J, Zhou W, Han N, Pang XM, et al. BH3-only proteins target BCL-xL/MCL-1, not BAX/BAK, to initiate apoptosis. Cell Res. 2019;29:942–52.
Ku B, Liang C, Jung JU, Oh BH. Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res. 2011;21:627–41.
Simmons MJ, Fan G, Zong WX, Degenhardt K, White E, Gélinas C. Bfl-1/A1 functions, similar to Mcl-1, as a selective tBid and Bak antagonist. Oncogene. 2008;27:1421–8.
Yan W, Samson M, Jégou B, Toppari J. Bcl-w forms complexes with Bax and Bak, and elevated ratios of Bax/Bcl-w and Bak/Bcl-w correspond to spermatogonial and spermatocyte apoptosis in the testis. Mol Endocrinol. 2000;14:682–99.
Moldoveanu T, Follis AV, Kriwacki RW, Green DR. Many players in BCL-2 family affairs. Trends Biochem Sci. 2014;39:101–11.
Chittenden T, Flemington C, Houghton AB, Ebb RG, Gallo GJ, Elangovan B, et al. A conserved domain in Bak, distinct from BH1 and BH2, mediates cell death and protein binding functions. EMBO J. 1995;14:5589–96.
Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–6.
Lee EF, Grabow S, Chappaz S, Dewson G, Hockings C, Kluck RM, et al. Physiological restraint of Bak by Bcl-xL is essential for cell survival. Genes Dev. 2016;30:1240–50.
Liu QA, Gehring K. Heterodimerization of BAK and MCL-1 activated by detergent micelles. J Biol Chem. 2010;285:41202–10.
Wang H, Guo M, Wei H, Chen Y. Targeting MCL-1 in cancer: current status and perspectives. J Hematol Oncol. 2021;14:67.
Wei AH, Roberts AW, Spencer A, Rosenberg AS, Siegel D, Walter RB, et al. Targeting MCL-1 in hematologic malignancies: rationale and progress. Blood Rev. 2020;44:100672.
Kehr S, Vogler M. It’s time to die: BH3 mimetics in solid tumors. Biochim Biophys Acta Mol Cell Res. 2021;1868:118987.
Senichkin VV, Streletskaia AY, Zhivotovsky B, Kopeina GS. Molecular comprehension of Mcl-1: from gene structure to cancer therapy. Trends Cell Biol. 2019;29:549–62.
Timucin AC, Basaga H, Kutuk O. Selective targeting of antiapoptotic BCL-2 proteins in cancer. Med Res Rev. 2019;39:146–75.
Diepstraten ST, Anderson MA, Czabotar PE, Lessene G, Strasser A, Kelly GL. The manipulation of apoptosis for cancer therapy using BH3-mimetic drugs. Nature Rev Cancer. 2022;22:45–64.
Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–8.
Merino D, Kelly GL, Lessene G, Wei AH, Roberts AW, Strasser A. BH3-Mimetic drugs: blazing the trail for new cancer medicines. Cancer Cell. 2018;34:879–91.
Liu JJ, Yin FK, Wang ZQ, Song T, Zhang ZC. An updated patent review of Mcl-1 inhibitors (2020-2022). Expert Opin Ther Pat. 2023;33:371–83.
Caenepeel S, Brown SP, Belmontes B, Moody G, Keegan KS, Chui D, et al. AMG 176, a selective MCL1 inhibitor, is effective in hematologic cancer models alone and in combination with established therapies. Cancer Discov. 2018;8:1582–97.
Dai HM, Ding HS, Meng XW, Peterson KL, Schneider PA, Karp JE, et al. Constitutive BAK activation as a determinant of drug sensitivity in malignant lymphohematopoietic cells. Genes Dev. 2015;29:2140–52.
Liu DY, Hou XN, Wu WY, Zafagnin V, Li YJ, Correia C, et al. Constitutive BAK/MCL1 complexes predict paclitaxel and S63845 sensitivity of ovarian cancer (vol 12, 1002, 2021). Cell Death Disease. 2021;12:1002.
Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538:477–82.
Yuda J, Will C, Phillips DC, Abraham L, Alvey C, Avigdor A, et al. Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients. Commun Med. 2023;3:154.
Tron AE, Belmonte MA, Adam A, Aquila BM, Boise LH, Chiarparin E, et al. Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia. Nature Commun. 2018;9:5341.
Wang HL, Guo M, Wei HD, Chen YH. Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins. Comput Struct Biotec. 2023;21:3760–7.
Fleischer B, Schulze-Bergkamen H, Schuchmann M, Weber A, Biesterfeld S, Muller M, et al. Mcl-1 is an anti-apoptotic factor for human hepatocellular carcinoma. Int J Oncol. 2006;28:25–32.
Michalski M, Bauer M, Walz F, Tumen D, Heumann P, Stockert P, et al. Simultaneous inhibition of Mcl-1 and Bcl-2 induces synergistic cell death in hepatocellular carcinoma. Biomedicines. 2023;11:1666.
Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–65.
Murray JB, Davidson J, Chen I, Davis B, Dokurno P, Graham CJ, et al. Establishing drug discovery and identification of hit series for the anti-apoptotic proteins, Bcl-2 and Mcl-1. ACS Omega. 2019;4:8892–906.
Fire E, Gullá SV, Grant RA, Keating AE. Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes. Protein Sci. 2010;19:507–19.
Czabotar PE, Lee EF, van Delft MF, Day CL, Smith BJ, Huang DC, et al. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc Natl Acad Sci USA. 2007;104:6217–22.
Dutta S, Gullá S, Chen TS, Fire E, Grant RA, Keating AE. Determinants of BH3 binding specificity for Mcl-1 versus BcI-x. J Mol Biol. 2010;398:747–62.
Denis C, Santos JSD, Bureau R, Voisin-Chiret AS. Hot-Spots of Mcl-1 protein. J Med Chem. 2020;63:928–43.
Merino D, Whittle JR, Vaillant F, Serrano A, Gong JN, Giner G, et al. Synergistic action of the MCL-1 inhibitor S63845 with current therapies in preclinical models of triple-negative and HER2-amplified breast cancer. Sci Transl Med. 2017;9:eaam7049.
Kehr S, Haydn T, Bierbrauer A, Irmer B, Vogler M, Fulda S. Targeting BCL-2 proteins in pediatric cancer: Dual inhibition of BCL-XL and MCL-1 leads to rapid induction of intrinsic apoptosis. Cancer Lett. 2020;482:19–32.
Senichkin VV, Pervushin NV, Zamaraev AV, Sazonova EV, Zuev AP, Streletskaia AY, et al. Bak and Bcl-xL participate in regulating sensitivity of solid tumor-derived cell lines to Mcl-1 inhibitors. Cancers. 2022;14:181.
Bruncko M, Wang L, Sheppard GS, Phillips DC, Tahir SK, Xue J, et al. Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity (vol 58, pg 2180, 2015). J Med Chem. 2015;58:4089–4089.
Bedoui S, Herold MJ, Strasser A. Emerging connectivity of programmed cell death pathways and its physiological implications. Nature Rev Mol Cell Biol. 2020;21:678–95.
Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403.
Zhai DY, Jin CF, Huang ZW, Satterthwait AC, Reed JC. Differential regulation of Bax and Bak by anti-apoptotic Bcl-2 family proteins Bcl-B and Mcl-1. J Biol Chem. 2008;283:9580–6.
Wolf P, Schoeniger A, Edlich F. Pro-apoptotic complexes of BAX and BAK on the outer mitochondrial membrane. Biochim Biophys Acta Mol Cell Res. 2022;1869:119317.
Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–54.
Iyer S, Bell F, Westphal D, Anwari K, Gulbis J, Smith BJ, et al. Bak apoptotic pores involve a flexible C-terminal region and juxtaposition of the C-terminal transmembrane domains. Cell Death Differ. 2015;22:1665–75.
Okamoto T, Zobel K, Fedorova A, Quan C, Yang H, Fairbrother WJ, et al. Stabilizing the pro-apoptotic BimBH3 helix (BimSAHB) does not necessarily enhance affinity or biological activity. ACS Chem Biol. 2013;8:297–302.
Czabotar PE, Lee EF, Thompson GV, Wardak AZ, Fairlie WD, Colman PM. Mutation to Bax beyond the BH3 domain disrupts interactions with pro-survival proteins and promotes apoptosis. J Biol Chem. 2011;286:7123–31.
Wei G, Margolin AA, Haery L, Brown E, Cucolo L, Julian B, et al. Chemical genomics identifies small-molecule repressors and BCL-xL as a predictor of MCL1 dependency. Cancer Cell. 2012;21:547–62.
Szlavik Z, Csekei M, Paczal A, Szabo ZB, Sipos S, Radics G, et al. Discovery of S64315, a potent and selective Mcl-1 inhibitor. J Med Chem. 2020;63:13762–95.
Nikolovska-Coleska Z, Wang RX, Fang XL, Pan HG, Tomita Y, Li P, et al. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332:261–73.
Clifton MC, Dranow DM, Leed A, Fulroth B, Fairman JW, Abendroth J, et al. A maltose-binding protein fusion construct yields a robust crystallography platform for MCL1. PLoS ONE. 2015;10:e0125010.
Acknowledgements
We thank the staff at BL17B1/BL18U1/BL19U1 beamlines at SSRF of the National Facility for Protein Science in Shanghai (NFPS), Shanghai Advanced Research Institute, Chinese Academy of Sciences, for providing technical support in X-ray diffraction data collection and analysis. We thank Department of Clinical Pharmacology, Xiangya Hospital, Central South University for providing Biacore 8 K instrument and technical support.
Funding
The present study was financially supported by the Natural Science Foundation of China (82470176, 82273496, 31900880 and 82172654), Hunan Provincial Science and Technology Department (2021RC4012), Changjiang Scholars Award Program of Ministry of Education, Natural Science Foundation of Hunan Province (2023JJ20092 and 2023JJ30863) and Central South University Innovation-Driven Research Program (2023CXQD076).
Author information
Authors and Affiliations
Contributions
YC and Wei H conceived the project. MG, Wang H, SX, LQ, JW, and Wei H performed experiments. M.G. and Wang H performed data collection and structure determination. XC and XL helped with data analysis. MG, XL, YC, and Wei H prepared the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
This study was approved by the ethics committee of the affiliated Xiangya Hospital of Central South University. All methods were performed in accordance with the relevant guidelines and regulations. Human cell lines were treated as described in the methods section of this manuscript. This study did not involve human subjects research or experiments on live vertebrates.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wei, H., Wang, H., Xiang, S. et al. Deciphering molecular specificity in MCL-1/BAK interaction and its implications for designing potent MCL-1 inhibitors. Cell Death Differ 32, 991–999 (2025). https://doi.org/10.1038/s41418-025-01454-2
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-025-01454-2
This article is cited by
-
CYP51A1 in health and disease: from sterol metabolism to regulated cell death
Cell Death Discovery (2025)