Abstract
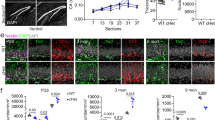
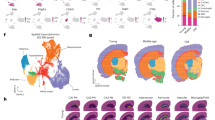
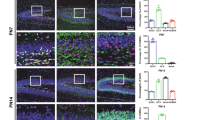
The dentate gyrus (DG), a crucial region of the hippocampus responsible for learning, spatial encoding, and memory formation, undergoes its main development and maturation after birth. Despite its importance, the regulatory mechanisms underlying postnatal DG development remain poorly understood. This study is aimed to investigate the role of H3 lysine 4 trimethylation (H3K4me3) and H3 lysine 27 trimethylation (H3K27me3) in the development and function of the postnatal DG. We show robust enrichment of H3K4me3 in the subgranular zone (SGZ), a primary neurogenic region, while high levels of H3K27me3 were mainly presented in granule cell layer. Enhanced H3K4me3 level facilitated proliferation and development of neonatal mouse neural stem cells (NSCs), promoted differentiation towards GABA neurons, as well as improved mouse spatial learning and memory. Enhancing H3K27me3 level exerts the opposite function, additionally promoting NSCs entry into a quiescent-like state. During the neuronal differentiation of NSCs, the integration of RNA-Seq and ChIP-Seq datasets reveals that H3K4me3 and H3K27me3 co-regulate the expression of genes essential for neural development, such as Gli1, through the formation of bivalent domains. Manipulation activation of the Shh/Gli1 pathway abolishes the effect of alterations in the levels of H3K4me3 and H3K27me3 in NSCs. Based on these findings, we propose that H3K4me3 and H3K27me3 serve as molecular “switches” to dynamically regulate NSCs proliferation and differentiation and in turn, influence the postnatal developmental progression of DG, additionally to provide potential therapeutic targets for treating diseases associated with abnormal hippocampal development.

During dentate gyrus development in neonatal mice, the active transcription mark H3K4me3 and the repressive mark H3K27me3 are co-localized at the promoter regions of essential neurodevelopmental genes, and thus forming bivalent chromatin domains in neural stem cells. These domains serve as a “molecular switch” that regulates the dynamic processes of cell proliferation and differentiation. The enhanced ratio of H3K4me3 to H3K27me3 markedly upregulates the expression related genes, thereby promoting cell proliferation and neuronal differentiation, ultimately leading to improved spatial learning and memory. Conversely, decreasing this ratio has the opposite effect.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The ChIP-seq and RNA-seq data have been deposited to NCBI database with the BioProject ID PRJNA 1179734 and are publicly available as of the date of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
Borzello M, Ramirez S, Treves A, Lee I, Scharfman H, Stark C, et al. Assessments of dentate gyrus function: discoveries and debates. Nat Rev Neurosci. 2023;24:502–17.
Tobin MK, Musaraca K, Disouky A, Shetti A, Bheri A, Honer WG, et al. Human hippocampal neurogenesis persists in aged adults and Alzheimer’s disease patients. Cell Stem Cell. 2019;24:974–82.e973.
Holland D, Chang L, Ernst TM, Curran M, Buchthal SD, Alicata D, et al. Structural growth trajectories and rates of change in the first 3 months of infant brain development. JAMA Neurol. 2014;71:1266–74.
Bond AM, Ming GL, Song H. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell. 2015;17:385–95.
Obernier K, Alvarez-Buylla A. Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development. 2019;146:dev156059.
Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017;49:e324.
Qian S, Lv X, Scheid RN, Lu L, Yang Z, Chen W, et al. Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL. Nat Commun. 2018;9:2425.
Park J, Lee K, Kim K, Yi SJ. The role of histone modifications: from neurodevelopment to neurodiseases. Signal Transduct Target Ther. 2022;7:217.
Shan Y, Zhang Y, Zhao Y, Wang T, Zhang J, Yao J, et al. JMJD3 and UTX determine fidelity and lineage specification of human neural progenitor cells. Nat Commun. 2020;11:382.
Shen E, Shulha H, Weng Z, Akbarian S. Regulation of histone H3K4 methylation in brain development and disease. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130514.
Zhang Z, Manaf A, Li Y, Perez SP, Suganthan R, Dahl JA, et al. Histone methylations define neural stem/progenitor cell subtypes in the mouse subventricular zone. Mol Neurobiol. 2020;57:997–1008.
Yang Z, Qian S, Scheid RN, Lu L, Chen X, Liu R, et al. EBS is a bivalent histone reader that regulates floral phase transition in Arabidopsis. Nat Genet. 2018;50:1247–53.
Dunican DS, Mjoseng HK, Duthie L, Flyamer IM, Bickmore WA, Meehan RR. Bivalent promoter hypermethylation in cancer is linked to the H327me3/H3K4me3 ratio in embryonic stem cells. BMC Biol. 2020;18:25.
Li F, Wan M, Zhang B, Peng Y, Zhou Y, Pi C, et al. Bivalent histone modifications and development. Curr Stem Cell Res Ther. 2018;13:83–90.
Kumar D, Cinghu S, Oldfield AJ, Yang P, Jothi R. Decoding the function of bivalent chromatin in development and cancer. Genome Res. 2021;31:2170–84.
Macrae TA, Fothergill-Robinson J, Ramalho-Santos M. Regulation, functions and transmission of bivalent chromatin during mammalian development. Nat Rev Mol Cell Biol. 2023;24:6–26.
Matlik K, Govek EE, Paul MR, Allis CD, Hatten ME. Histone bivalency regulates the timing of cerebellar granule cell development. Genes Dev. 2023;37:570–89.
Lajud N, Torner L. Early life stress and hippocampal neurogenesis in the neonate: sexual dimorphism, long term consequences and possible mediators. Front Mol Neurosci. 2015;8:3.
Pan MR, Hsu MC, Chen LT, Hung WC. Orchestration of H3K27 methylation: mechanisms and therapeutic implication. Cell Mol Life Sci. 2018;75:209–23.
Poreba E, Lesniewicz K, Durzynska J. Histone-lysine N-methyltransferase 2 (KMT2) complexes - a new perspective. Mutat Res Rev Mutat Res. 2022;790:108443.
Kidder BL, Hu G, Zhao K. KDM5B focuses H3K4 methylation near promoters and enhancers during embryonic stem cell self-renewal and differentiation. Genome Biol. 2014;15:R32.
Hua C, Chen J, Li S, Zhou J, Fu J, Sun W, et al. KDM6 demethylases and their roles in human cancers. Front Oncol. 2021;11:779918.
Urban N, Blomfield IM, Guillemot F. Quiescence of adult mammalian neural stem cells: a highly regulated rest. Neuron. 2019;104:834–48.
Marques-Torrejon MA, Williams CAC, Southgate B, Alfazema N, Clements MP, Garcia-Diaz C, et al. LRIG1 is a gatekeeper to exit from quiescence in adult neural stem cells. Nat Commun. 2021;12:2594.
Sueda R, Imayoshi I, Harima Y, Kageyama R. High Hes1 expression and resultant Ascl1 suppression regulate quiescent vs. active neural stem cells in the adult mouse brain. Genes Dev. 2019;33:511–23.
Berg DA, Su Y, Jimenez-Cyrus D, Patel A, Huang N, Morizet D, et al. A common embryonic origin of stem cells drives developmental and adult neurogenesis. Cell. 2019;177:654–68.e615.
Ribas VT, Costa MR. Gene manipulation strategies to identify molecular regulators of axon regeneration in the central nervous system. Front Cell Neurosci. 2017;11:231.
Cwetsch AW, Pinto B, Savardi A, Cancedda L. In vivo methods for acute modulation of gene expression in the central nervous system. Prog Neurobiol. 2018;168:69–85.
Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell. 2012;22:25–37.
Cho E, Mysliwiec MR, Carlson CD, Ansari A, Schwartz RJ, Lee Y. Cardiac-specific developmental and epigenetic functions of Jarid2 during embryonic development. J Biol Chem. 2018;293:11659–73.
dal Maschio M, Ghezzi D, Bony G, Alabastri A, Deidda G, Brondi M, et al. High-performance and site-directed in utero electroporation by a triple-electrode probe. Nat Commun. 2012;3:960.
Caramello A, Galichet C, Rizzoti K, Lovell-Badge R. Dentate gyrus development requires a cortical hem-derived astrocytic scaffold. Elife. 2021;10:e63904.
Jimenez-Cyrus D, Adusumilli VS, Stempel MH, Maday S, Ming GL, Song H, et al. Molecular cascade reveals sequential milestones underlying hippocampal neural stem cell development into an adult state. Cell Rep. 2024;43:114339.
Carretero-Guillen A, Trevino M, Gomez-Climent MA, Dogbevia GK, Bertocchi I, Sprengel R, et al. Dentate gyrus is needed for memory retrieval. Mol Psychiatry. 2024;29:2939–50.
Hainmueller T, Bartos M. Dentate gyrus circuits for encoding, retrieval and discrimination of episodic memories. Nat Rev Neurosci. 2020;21:153–68.
Fanselow MS, Dong H-W. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron. 2010;65:7–19.
Blanco E, Gonzalez-Ramirez M, Alcaine-Colet A, Aranda S, Di Croce L. The bivalent genome: characterization, structure, and regulation. Trends Genet. 2020;36:118–31.
Cai C, Hu X, Dai P, Zhang T, Jiang M, Wang L, et al. c-Myc regulates neural stem cell quiescence and activation by coordinating the cell cycle and mitochondrial remodeling. Signal Transduct Target Ther. 2021;6:306.
Kerosuo L, Neppala P, Hsin J, Mohlin S, Vieceli FM, Torok Z, et al. Enhanced expression of MycN/CIP2A drives neural crest toward a neural stem cell-like fate: Implications for priming of neuroblastoma. Proc Natl Acad Sci USA. 2018;115:E7351–E7360.
Johnson CA, Ghashghaei HT. Sp2 regulates late neurogenic but not early expansive divisions of neural stem cells underlying population growth in the mouse cortex. Development. 2020;147:dev186056.
Samanta J, Grund EM, Silva HM, Lafaille JJ, Fishell G, Salzer JL. Inhibition of Gli1 mobilizes endogenous neural stem cells for remyelination. Nature. 2015;526:448–52.
Fong BC, Chakroun I, Iqbal MA, Paul S, Bastasic J, O’Neil D, et al. The Rb/E2F axis is a key regulator of the molecular signatures instructing the quiescent and activated adult neural stem cell state. Cell Rep. 2022;41:111578.
Jacobs CT, Huang P. Complex crosstalk of Notch and Hedgehog signalling during the development of the central nervous system. Cell Mol Life Sci. 2021;78:635–44.
Yamaguchi M, Seki T, Imayoshi I, Tamamaki N, Hayashi Y, Tatebayashi Y, et al. Neural stem cells and neuro/gliogenesis in the central nervous system: understanding the structural and functional plasticity of the developing, mature, and diseased brain. J Physiol Sci. 2016;66:197–206.
Dong J, Pan YB, Wu XR, He LN, Liu XD, Feng DF, et al. A neuronal molecular switch through cell-cell contact that regulates quiescent neural stem cells. Sci Adv. 2019;5:eaav4416.
Yao J, Dai S, Zhu R, Tan J, Zhao Q, Yin Y, et al. Deciphering molecular heterogeneity and dynamics of human hippocampal neural stem cells at different ages and injury states. Elife. 2024;12:RP89507.
Zhao T, Hong Y, Yan B, Huang S, Ming GL, Song H. Epigenetic maintenance of adult neural stem cell quiescence in the mouse hippocampus via Setd1a. Nat Commun. 2024;15:5674.
Meshorer E, Plath K. Chromatin and nuclear architecture in stem cells. Stem Cell Reports. 2020;15:1155–7.
Mu M, Li X, Dong L, Wang J, Cai Q, Hu Y, et al. METTL14 regulates chromatin bivalent domains in mouse embryonic stem cells. Cell Rep. 2023;42:113116.
Zhao C, Dong C, Frah M, Deng Y, Marie C, Zhang F, et al. Dual requirement of CHD8 for chromatin landscape establishment and histone methyltransferase recruitment to promote CNS myelination and repair. Dev Cell. 2018;45:753–68.e758.
Nguyen H, Kerimoglu C, Pirouz M, Pham L, Kiszka KA, Sokpor G, et al. Epigenetic regulation by BAF complexes limits neural stem cell proliferation by suppressing Wnt signaling in late embryonic development. Stem Cell Reports. 2018;10:1734–50.
Hatch SB, Yapp C, Montenegro RC, Savitsky P, Gamble V, Tumber A, et al. Assessing histone demethylase inhibitors in cells: lessons learned. Epigenetics Chromatin. 2017;10:9.
Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–8.
Coulter DA, Carlson GC. Functional regulation of the dentate gyrus by GABA-mediated inhibition. Prog Brain Res. 2007;163:235–43.
Schmitz TW, Correia MM, Ferreira CS, Prescot AP, Anderson MC. Hippocampal GABA enables inhibitory control over unwanted thoughts. Nat Commun. 2017;8:1311.
Stefanelli T, Bertollini C, Luscher C, Muller D, Mendez P. Hippocampal somatostatin interneurons control the size of neuronal memory ensembles. Neuron. 2016;89:1074–85.
Tzilivaki A, Tukker JJ, Maier N, Poirazi P, Sammons RP, Schmitz D. Hippocampal GABAergic interneurons and memory. Neuron. 2023;111:3154–75.
Hosp JA, Struber M, Yanagawa Y, Obata K, Vida I, Jonas P, et al. Morpho-physiological criteria divide dentate gyrus interneurons into classes. Hippocampus. 2014;24:189–203.
Pelkey KA, Chittajallu R, Craig MT, Tricoire L, Wester JC, McBain CJ. Hippocampal GABAergic inhibitory interneurons. Physiol Rev. 2017;97:1619–747.
Morris DR, Levenson CW. Zinc regulation of transcriptional activity during retinoic acid-induced neuronal differentiation. J Nutr Biochem. 2013;24:1940–4.
Yao PJ, Petralia RS, Mattson MP. Sonic Hedgehog signaling and hippocampal neuroplasticity. Trends Neurosci. 2016;39:840–50.
Favaro R, Valotta M, Ferri AL, Latorre E, Mariani J, Giachino C, et al. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci. 2009;12:1248–56.
Stecca B, Ruiz i Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J. 2009;28:663–76.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (No.81901156, 82271200, 82400174, 82171308); Research Council of Norway through its Centers of Excellence scheme (332713).
Author information
Authors and Affiliations
Contributions
YL and HZ performed all experiments and data processing. YL and HY performed cell culture and immunostaining. WL performed immunostaining and confocal imaging. JZ performed ChIP experiments. KM and XZ performed animal behavior analysis. CH, XC, and HL helped designed and refine the experiments. AK and ZZ conceived the project and designed the experiments, with assistance from MB. ZZ wrote the manuscript, with input and editing from MB and AK.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics declarations
All methods were performed in accordance with the relevant guidelines and regulations. Mouse experiments were approved by the Institutional Animal Care and Use Committee of Xi’an Jiaotong University (No. XJTUAE2022-445). Mice were housed under standard conditions, which entailed housing in facilities with a 12-h light/dark cycle, ad libitum access to food and water, and a temperature range of 20–24 °C. Stringent measures were taken to mitigate animal suffering and reduce the overall number of animals utilized. We thank the Laboratory Animal Center at Xi’an Jiaotong University for their assistance with housing and breeding experimental mice. This study does not directly involve human subjects or human data that requires ethical approval.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Luan, Y., Zhang, H., Liu, Y. et al. Inverse and dynamic levels of H3K4me3 and H3K27me3 regulate mouse postnatal dental gyrus development. Cell Death Differ 33, 219–235 (2026). https://doi.org/10.1038/s41418-025-01563-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-025-01563-y