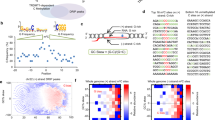
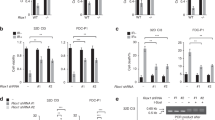
Abstract
Repair of DNA double-strand breaks (DSBs) is essential for cells to maintain genome stability and cell survival. While cyclic GMP-AMP synthase (cGAS) is best known for its role in innate immunity, emerging evidence reveals that it plays regulatory roles in DNA damage response. In this study, we demonstrate that cGAS suppresses PARP1-mediated poly(ADP-ribose) (PAR) formation at both double-stranded DNA (dsDNA) and DNA:RNA hybrids. As a result, cGAS deficiency enhances PARP1-mediated microhomology-mediated end joining (MMEJ). Since MMEJ competes with transcription-coupled homologous recombination (TC-HR) at actively transcribed genomic regions, cGAS-mediated suppression of MMEJ consequently leads to efficient TC-HR with increased recruitment of RAD52 and RAD51. Mechanistically, the zinc finger domain of cGAS binds PAR, inhibiting PARP1 activation. In prostate cancer cells, loss of cGAS increases dependency on PARP1, rendering them more sensitive to PARP inhibitors. Collectively, our findings uncover a noncanonical role for cGAS in negatively regulating PARP1-mediated MMEJ and suggest its potential as a therapeutic biomarker in prostate cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data is contained within the manuscript and/or supplementary files. The original western blot images are provided in the Supplementary File.
References
Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends cell Biol. 2016;26:52–64.
Schwer B, Wei PC, Chang AN, Kao J, Du Z, Meyers RM, et al. Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proc Natl Acad Sci USA. 2016;113:2258–63.
Wei PC, Chang AN, Kao J, Du Z, Meyers RM, Alt FW, et al. Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell. 2016;164:644–55.
Marnef A, Cohen S, Legube G. Transcription-coupled DNA double-strand break repair: active genes need special care. J Mol Biol. 2017;429:1277–88.
Yang H, Lachtara EM, Ran X, Hopkins J, Patel PS, Zhu X, et al. The RNA m5C modification in R-loops as an off switch of Alt-NHEJ. Nat Commun. 2023;14:6114.
Yang H, Lan L. Transcription-coupled DNA repair protects genome stability upon oxidative stress-derived DNA strand breaks. FEBS Lett. 2024.
Marnef A, Legube G. R-loops as Janus-faced modulators of DNA repair. Nat cell Biol. 2021;23:305–13.
Petermann E, Lan L, Zou L. Sources, resolution and physiological relevance of R-loops and RNA-DNA hybrids. Nat Rev Mol cell Biol. 2022;23:521–40.
Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:366–74.
Wei L, Nakajima S, Bohm S, Bernstein KA, Shen Z, Tsang M, et al. DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc Natl Acad Sci USA. 2015;112:E3495–3504.
Ouyang J, Yadav T, Zhang JM, Yang H, Rheinbay E, Guo H, et al. RNA transcripts stimulate homologous recombination by forming DR-loops. Nature. 2021;594:283–8.
Teng Y, Yadav T, Duan M, Tan J, Xiang Y, Gao B, et al. ROS-induced R loops trigger a transcription-coupled but BRCA1/2-independent homologous recombination pathway through CSB. Nat Commun. 2018;9:4115.
Chen H, Yang H, Zhu X, Yadav T, Ouyang J, Truesdell SS, et al. m(5)C modification of mRNA serves a DNA damage code to promote homologous recombination. Nat Commun. 2020;11:2834.
Sallmyr A, Tomkinson AE. Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J Biol Chem. 2018;293:10536–46.
Liu Q, Palomero L, Moore J, Guix I, Espin R, Aytes A, et al. Loss of TGFbeta signaling increases alternative end-joining DNA repair that sensitizes to genotoxic therapies across cancer types. Sci Transl Med. 2021, 13.
Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic acids Res. 2006;34:6170–82.
Patterson-Fortin J, D’Andrea AD. Exploiting the microhomology-mediated end-joining pathway in cancer therapy. Cancer Res. 2020;80:4593–4600.
Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91.
Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8.
Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–6.
Jiang H, Xue X, Panda S, Kawale A, Hooy RM, Liang F, et al. Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO J. 2019;38:e102718.
Zhen Z, Chen Y, Wang H, Tang H, Zhang H, Liu H, et al. Nuclear cGAS restricts L1 retrotransposition by promoting TRIM41-mediated ORF2p ubiquitination and degradation. Nat Commun. 2023;14:8217.
Chen H, Chen H, Zhang J, Wang Y, Simoneau A, Yang H, et al. cGAS suppresses genomic instability as a decelerator of replication forks. Sci Adv. 2020, 6.
Gentili M, Lahaye X, Nadalin F, Nader GPF, Puig Lombardi E, Herve S, et al. The N-terminal domain of cGAS determines preferential association with centromeric DNA and innate immune activation in the nucleus. Cell Rep. 2019;26:2377–2393 e2313.
Luedeman ME, Stroik S, Feng W, Luthman AJ, Gupta GP, Ramsden DA. Poly(ADP) ribose polymerase promotes DNA polymerase theta-mediated end joining by activation of end resection. Nat Commun. 2022;13:4547.
Laspata N, Kaur P, Mersaoui SY, Muoio D, Liu ZS, Bannister MH, et al. PARP1 associates with R-loops to promote their resolution and genome stability. Nucleic acids Res. 2023;51:2215–37.
Lan L, Nakajima S, Wei L, Sun L, Hsieh CL, Sobol RW, et al. Novel method for site-specific induction of oxidative DNA damage reveals differences in recruitment of repair proteins to heterochromatin and euchromatin. Nucleic acids Res. 2014;42:2330–45.
Houl JH, Ye Z, Brosey CA, Balapiti-Modarage LPF, Namjoshi S, Bacolla A, et al. Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death. Nat Commun. 2019;10:5654.
Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85.
Kamaletdinova T, Fanaei-Kahrani Z, Wang ZQ. The enigmatic function of PARP1: from PARylation activity to PAR readers. Cells. 2019, 8.
Wang F, Zhao M, Chang B, Zhou Y, Wu X, Ma M, et al. Cytoplasmic PARP1 links the genome instability to the inhibition of antiviral immunity through PARylating cGAS. Mol cell. 2022;82:2032–2049 e2037.
Verhagen PC, Hermans KG, Brok MO, van Weerden WM, Tilanus MG, de Weger RA, et al. Deletion of chromosomal region 6q14-16 in prostate cancer. Int J cancer. 2002;102:142–7.
Kluth M, Jung S, Habib O, Eshagzaiy M, Heinl A, Amschler N, et al. Deletion lengthening at chromosomes 6q and 16q targets multiple tumor suppressor genes and is associated with an increasingly poor prognosis in prostate cancer. Oncotarget. 2017;8:108923–35.
Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304 e296.
Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci USA. 2019;116:11428–36.
Tsujino T, Takai T, Hinohara K, Gui F, Tsutsumi T, Bai X, et al. CRISPR screens reveal genetic determinants of PARP inhibitor sensitivity and resistance in prostate cancer. Nat Commun. 2023;14:252.
Zhang H, Jiang L, Du X, Qian Z, Wu G, Jiang Y, et al. The cGAS-Ku80 complex regulates the balance between two end joining subpathways. Cell Death Differ. 2024;31:792–803.
Barnett KC, Coronas-Serna JM, Zhou W, Ernandes MJ, Cao A, Kranzusch PJ, et al. Phosphoinositide interactions position cGAS at the plasma membrane to ensure efficient distinction between self- and viral DNA. Cell. 2019;176:1432–1446 e1411.
Yang H, Lan L. Immunostaining and protein pull-down of methyl-5-cytosine-marked RNA:DNA hybrids. Methods Mol Biol. 2022;2528:271–6.
Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA, Zou L. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol cell. 2017;65:832–847 e834.
Acknowledgements
This work was supported by grants from National Institutes of Health R01CA262524 (LJ), R01CA279410 (LJ), R01CA282939 (LL); Department of Defense W81XWH-22-1-0477 (LJ).
Author information
Authors and Affiliations
Contributions
HY conducted the major experiments and data analysis. BG, BW, WL, XZ, LP, JL, and FG participated in the experiments and data analysis. WZ participated in experimental design and data analysis. LJ and LL provided overall experimental guidance. All authors acknowledge their specific contributions and have reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
Ethics approval was not required for this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, H., Gao, B., Wu, B. et al. cGAS restricts PARP1-mediated microhomology-mediated end joining by suppressing poly-ADP-ribosylation. Cell Death Differ (2025). https://doi.org/10.1038/s41418-025-01637-x
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41418-025-01637-x